Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity
- PMID: 16314439
- PMCID: PMC2213337
- DOI: 10.1084/jem.20051625
Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity
Abstract
Sulfatide derived from the myelin stimulates a distinct population of CD1d-restricted natural killer T (NKT) cells. Cis-tetracosenoyl sulfatide is one of the immunodominant species in myelin as identified by proliferation, cytokine secretion, and CD1d tetramer staining. The crystal structure of mouse CD1d in complex with cis-tetracosenoyl sulfatide at 1.9 A resolution reveals that the longer cis-tetracosenoyl fatty acid chain fully occupies the A' pocket of the CD1d binding groove, whereas the sphingosine chain fills up the F' pocket. A precise hydrogen bond network in the center of the binding groove orients and positions the ceramide backbone for insertion of the lipid tails in their respective pockets. The 3'-sulfated galactose headgroup is highly exposed for presentation to the T cell receptor and projects up and away from the binding pocket due to its beta linkage, compared with the more intimate binding of the alpha-glactosyl ceramide headgroup to CD1d. These structure and binding data on sulfatide presentation by CD1d have important implications for the design of therapeutics that target T cells reactive for myelin glycolipids in autoimmune diseases of the central nervous system.
Figures



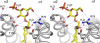

References
-
- Bendelac, A. 1995. Mouse NK1+ T cells. Curr. Opin. Immunol. 7:367–374. - PubMed
-
- Wilson, S.B., and T.L. Delovitch. 2003. Janus-like role of regulatory iNKT cells in autoimmune disease and tumour immunity. Nat. Rev. Immunol. 3:211–222. - PubMed
-
- Kawano, T., J. Cui, Y. Koezuka, I. Toura, Y. Kaneko, K. Motoki, H. Ueno, R. Nakagawa, H. Sato, E. Kondo, et al. 1997. CD1d-restricted and TCR-mediated activation of vα14 NKT cells by glycosylceramides. Science. 278:1626–1629. - PubMed
-
- Zeng, Z., A.R. Castano, B.W. Segelke, E.A. Stura, P.A. Peterson, and I.A. Wilson. 1997. Crystal structure of mouse CD1: an MHC-like fold with a large hydrophobic binding groove. Science. 277:339–345. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

