The ternary complex factor Net regulates cell migration through inhibition of PAI-1 expression
- PMID: 16314510
- PMCID: PMC1316955
- DOI: 10.1128/MCB.25.24.10853-10862.2005
The ternary complex factor Net regulates cell migration through inhibition of PAI-1 expression
Abstract
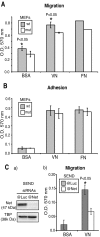
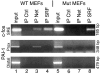
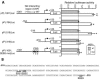
Net, Elk-1, and Sap-1 are members of the ternary complex factor (TCF) subfamily of Ets transcription factors. They form ternary complexes with serum response factor (SRF) on serum response elements of immediate early genes such as c-fos and egr-1 and mediate responses to growth factors and mitogen-activated protein kinase signaling. Although the TCFs have been extensively studied as intermediates in signaling cascades, surprisingly little is known about their different target genes and physiological functions. We report that Net homozygous mutant mouse embryonic fibroblasts have a defect in cell migration. This defect results at least in part from increased expression of plasminogen activator inhibitor type 1 (PAI-1), a serine protease inhibitor (serpin) that controls extracellular proteolysis and cell matrix adhesion. The defect in cell migration can be reverted by the addition of a PAI-1 blocking antibody. Net represses PAI-1 promoter activity and binds to a specific region of the promoter containing Ets binding sites in the absence of SRF. We conclude that Net is a negative regulator of PAI-1 expression and is thereby involved in cell migration.
Figures




Similar articles
-
The ternary complex factor net is downregulated by hypoxia and regulates hypoxia-responsive genes.Mol Cell Biol. 2007 Jun;27(11):4133-41. doi: 10.1128/MCB.01867-06. Epub 2007 Apr 2. Mol Cell Biol. 2007. PMID: 17403894 Free PMC article.
-
Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex.J Mol Biol. 2001 Nov 30;314(3):495-506. doi: 10.1006/jmbi.2001.5138. J Mol Biol. 2001. PMID: 11846562
-
Ternary complex factors SAP-1 and Elk-1, but not net, are functionally equivalent in thymocyte development.J Immunol. 2010 Jul 15;185(2):1082-92. doi: 10.4049/jimmunol.1000472. Epub 2010 Jun 16. J Immunol. 2010. PMID: 20554967
-
Transcriptional regulation of the plasminogen activator inhibitor type 1--with an emphasis on negative regulation.Thromb Haemost. 2008 Dec;100(6):1007-13. Thromb Haemost. 2008. PMID: 19132223 Review.
-
Serum response factor: discovery, biochemistry, biological roles and implications for tissue injury healing.J Physiol Pharmacol. 2002 Jun;53(2):147-57. J Physiol Pharmacol. 2002. PMID: 12120892 Review.
Cited by
-
The ternary complex factor net is downregulated by hypoxia and regulates hypoxia-responsive genes.Mol Cell Biol. 2007 Jun;27(11):4133-41. doi: 10.1128/MCB.01867-06. Epub 2007 Apr 2. Mol Cell Biol. 2007. PMID: 17403894 Free PMC article.
-
Involvement of net and Hif1alpha in distinct yet intricately linked hypoxia-induced signaling pathways.J Biol Chem. 2010 Jul 9;285(28):21223-32. doi: 10.1074/jbc.M110.121723. Epub 2010 Apr 28. J Biol Chem. 2010. PMID: 20427288 Free PMC article.
-
ELK3 Expression Correlates With Cell Migration, Invasion, and Membrane Type 1-Matrix Metalloproteinase Expression in MDA-MB-231 Breast Cancer Cells.Gene Expr. 2015;16(4):197-203. doi: 10.3727/105221615X14399878166276. Gene Expr. 2015. PMID: 26637400 Free PMC article.
-
Net expression inhibits the growth of pancreatic ductal adenocarcinoma cell PL45 in vitro and in vivo.PLoS One. 2013;8(2):e57818. doi: 10.1371/journal.pone.0057818. Epub 2013 Feb 28. PLoS One. 2013. Retraction in: PLoS One. 2024 Dec 12;19(12):e0315984. doi: 10.1371/journal.pone.0315984. PMID: 23469073 Free PMC article. Retracted.
-
SRF Co-factors Control the Balance between Cell Proliferation and Contractility.Mol Cell. 2016 Dec 15;64(6):1048-1061. doi: 10.1016/j.molcel.2016.10.016. Epub 2016 Nov 17. Mol Cell. 2016. PMID: 27867007 Free PMC article.
References
-
- Ayadi, A., M. Suelves, P. Dolle, and B. Wasylyk. 2001. Net, an Ets ternary complex transcription factor, is expressed in sites of vasculogenesis, angiogenesis, and chondrogenesis during mouse development. Mech. Dev. 102:205-208. - PubMed
-
- Bajou, K., C. Maillard, M. Jost, R. H. Lijnen, A. Gils, P. Declerck, P. Carmeliet, J. M. Foidart, and A. Noel. 2004. Host-derived plasminogen activator inhibitor-1 (PAI-1) concentration is critical for in vivo tumoral angiogenesis and growth. Oncogene 23:6986-6990. - PubMed
-
- Bass, R., and V. Ellis. 2002. Cellular mechanisms regulating non-haemostatic plasmin generation. Biochem. Soc. Trans. 30:189-194. - PubMed
-
- Bauer, S., J. Pollheimer, J. Hartmann, P. Husslein, J. D. Aplin, and M. Knofler. 2004. Tumor necrosis factor-alpha inhibits trophoblast migration through elevation of plasminogen activator inhibitor-1 in first-trimester villous explant cultures. J. Clin. Endocrinol. Metab. 89:812-822. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
