Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein
- PMID: 16319927
- PMCID: PMC1356327
- DOI: 10.1038/sj.emboj.7600886
Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein
Abstract
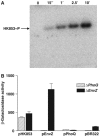
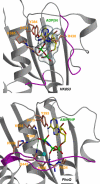
The large majority of histidine kinases (HKs) are multifunctional enzymes having autokinase, phosphotransfer and phosphatase activities, and most of these are transmembrane sensor proteins. Sensor HKs possess conserved cytoplasmic phosphorylation and ATP-binding kinase domains. The different enzymatic activities require participation by one or both of these domains, implying the need for different conformational states. The catalytic domains are linked to the membrane through a coiled-coil segment that sometimes includes other domains. We describe here the first crystal structure of the complete cytoplasmic region of a sensor HK, one from the thermophile Thermotoga maritima in complex with ADPbetaN at 1.9 A resolution. The structure reveals previously unidentified functions for several conserved residues and reveals the relative disposition of domains in a state seemingly poised for phosphotransfer. The structure thereby inspires hypotheses for the mechanisms of autophosphorylation, phosphotransfer and response-regulator dephosphorylation, and for signal transduction through the coiled-coil segment. Mutational tests support the functional relevance of interdomain contacts.
Figures

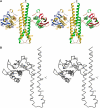

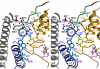


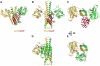
References
-
- Aravind L, Ponting CP (1999) The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett 176: 111–116 - PubMed
-
- Bilwes AM, Alex LA, Crane BR, Simon MI (1999) Structure of CheA, a signal-transducing histidine kinase. Cell 96: 131–141 - PubMed
-
- Bilwes AM, Quezada CM, Croal LR, Crane BR, Simon MI (2001) Nucleotide binding by the histidine kinase CheA. Nat Struct Biol 8: 353–360 - PubMed
-
- Birck C, Mourey L, Gouet P, Fabry B, Schumacher J, Rousseau P, Kahn D, Samama JP (1999) Conformational changes induced by phosphorylation of the FixJ receiver domain. Struct Fold Des 7: 1505–1515 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

