The ClgR protein regulates transcription of the clpP operon in Bifidobacterium breve UCC 2003
- PMID: 16321946
- PMCID: PMC1317013
- DOI: 10.1128/JB.187.24.8411-8426.2005
The ClgR protein regulates transcription of the clpP operon in Bifidobacterium breve UCC 2003
Abstract
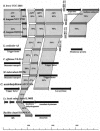
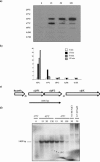
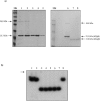
Five clp genes (clpC, clpB, clpP1, clpP2, and clpX), representing chaperone- and protease-encoding genes, were previously identified in Bifidobacterium breve UCC 2003. In the present study, we characterize the B. breve UCC 2003 clpP locus, which consists of two paralogous genes, designated clpP1 and clpP2, whose deduced protein products display significant similarity to characterized ClpP peptidases. Transcriptional analyses showed that the clpP1 and clpP2 genes are transcribed in response to moderate heat shock as a bicistronic unit with a single promoter. The role of a clgR homologue, known to control the regulation of clpP gene expression in Streptomyces lividans and Corynebacterium glutamicum, was investigated by gel mobility shift assays and DNase I footprint experiments. We show that ClgR, which in its purified form appears to exist as a dimer, requires a proteinaceous cofactor to assist in specific binding to a 30-bp region of the clpP promoter region. In pull-down experiments, a 56-kDa protein copurified with ClgR, providing evidence that the two proteins also interact in vivo and that the copurified protein represents the cofactor required for ClgR activity. The prediction of the ClgR three-dimensional structure provides further insights into the binding mode of this protein to the clpP1 promoter region and highlights the key amino acid residues believed to be involved in the protein-DNA interaction.
Figures






Similar articles
-
The transcriptional activator ClgR controls transcription of genes involved in proteolysis and DNA repair in Corynebacterium glutamicum.Mol Microbiol. 2005 Jul;57(2):576-91. doi: 10.1111/j.1365-2958.2005.04710.x. Mol Microbiol. 2005. PMID: 15978086
-
clpC and clpP1P2 gene expression in Corynebacterium glutamicum is controlled by a regulatory network involving the transcriptional regulators ClgR and HspR as well as the ECF sigma factor sigmaH.Mol Microbiol. 2004 Apr;52(1):285-302. doi: 10.1111/j.1365-2958.2003.03979.x. Mol Microbiol. 2004. PMID: 15049827
-
Genetic and transcriptional organization of the clpC locus in Bifidobacterium breve UCC 2003.Appl Environ Microbiol. 2005 Oct;71(10):6282-91. doi: 10.1128/AEM.71.10.6282-6291.2005. Appl Environ Microbiol. 2005. PMID: 16204550 Free PMC article.
-
ClgR, a novel regulator of clp and lon expression in Streptomyces.J Bacteriol. 2004 May;186(10):3238-48. doi: 10.1128/JB.186.10.3238-3248.2004. J Bacteriol. 2004. PMID: 15126487 Free PMC article.
-
Regulation of the clpP1clpP2 operon by the pleiotropic regulator AdpA in Streptomyces lividans.Arch Microbiol. 2013 Dec;195(12):831-41. doi: 10.1007/s00203-013-0918-2. Epub 2013 Nov 7. Arch Microbiol. 2013. PMID: 24196782
Cited by
-
An interactive regulatory network controls stress response in Bifidobacterium breve UCC2003.J Bacteriol. 2009 Nov;191(22):7039-49. doi: 10.1128/JB.00897-09. Epub 2009 Sep 4. J Bacteriol. 2009. PMID: 19734308 Free PMC article.
-
Genome-Wide Assessment of Stress-Associated Genes in Bifidobacteria.Appl Environ Microbiol. 2022 Apr 12;88(7):e0225121. doi: 10.1128/aem.02251-21. Epub 2022 Mar 21. Appl Environ Microbiol. 2022. PMID: 35311508 Free PMC article.
-
Transposon mutagenesis in Bifidobacterium breve: construction and characterization of a Tn5 transposon mutant library for Bifidobacterium breve UCC2003.PLoS One. 2013 May 30;8(5):e64699. doi: 10.1371/journal.pone.0064699. Print 2013. PLoS One. 2013. PMID: 23737995 Free PMC article.
-
Degradation Mechanism of AAA+ Proteases and Regulation of Streptomyces Metabolism.Biomolecules. 2022 Dec 10;12(12):1848. doi: 10.3390/biom12121848. Biomolecules. 2022. PMID: 36551276 Free PMC article. Review.
-
Characterization of a Clp protease gene regulator and the reaeration response in Mycobacterium tuberculosis.PLoS One. 2010 Jul 16;5(7):e11622. doi: 10.1371/journal.pone.0011622. PLoS One. 2010. PMID: 20661284 Free PMC article.
References
-
- Bucca, G., A. M. E. Brassington, H. J. Schonfeld, and C. P. Smith. 2000. The HspR regulon of Streptomyces coelicolor: a role for the DnaK chaperone as a transcriptional co-repressor. Mol. Microbiol. 38:1093-1103. - PubMed
-
- Crecy-Lagard, V., P. Servant-Moisson, J. Viala, C. Grandvalet, and P. Mazodier. 1999. Alteration of the synthesis of the Clp ATP-dependent protease affects morphological and physiological differentiation in Streptomyces. Mol. Microbiol. 32:505-517. - PubMed
-
- Derre, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31:117-131. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

