Identification of a twin-arginine translocation system in Pseudomonas syringae pv. tomato DC3000 and its contribution to pathogenicity and fitness
- PMID: 16321949
- PMCID: PMC1317023
- DOI: 10.1128/JB.187.24.8450-8461.2005
Identification of a twin-arginine translocation system in Pseudomonas syringae pv. tomato DC3000 and its contribution to pathogenicity and fitness
Abstract
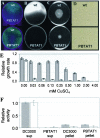
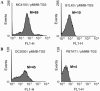
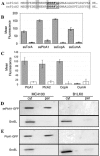
The bacterial plant pathogen Pseudomonas syringae pv. tomato DC3000 (DC3000) causes disease in Arabidopsis thaliana and tomato plants, and it elicits the hypersensitive response in nonhost plants such as Nicotiana tabacum and Nicotiana benthamiana. While these events chiefly depend upon the type III protein secretion system and the effector proteins that this system translocates into plant cells, additional factors have been shown to contribute to DC3000 virulence and still many others are likely to exist. Therefore, we explored the contribution of the twin-arginine translocation (Tat) system to the physiology of DC3000. We found that a tatC mutant strain of DC3000 displayed a number of phenotypes, including loss of motility on soft agar plates, deficiency in siderophore synthesis and iron acquisition, sensitivity to copper, loss of extracellular phospholipase activity, and attenuated virulence in host plant leaves. In the latter case, we provide evidence that decreased virulence of tatC mutants likely arises from a synergistic combination of (i) compromised fitness of bacteria in planta; (ii) decreased efficiency of type III translocation; and (iii) cytoplasmically retained virulence factors. Finally, we demonstrate a novel broad-host-range genetic reporter based on the green fluorescent protein for the identification of Tat-targeted secreted virulence factors that should be generally applicable to any gram-negative bacterium. Collectively, our evidence supports the notion that virulence of DC3000 is a multifactorial process and that the Tat system is an important virulence determinant of this phytopathogenic bacterium.
Figures




References
-
- Alfano, J. R., and A. Collmer. 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42:385-414. - PubMed
-
- Berks, B. C., T. Palmer, and F. Sargent. 2003. The Tat protein translocation pathway and its role in microbial physiology. Adv. Microb. Physiol. 47:187-254. - PubMed
-
- Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

