A genomic approach to the identification and characterization of HOXA13 functional binding elements
- PMID: 16321965
- PMCID: PMC1301594
- DOI: 10.1093/nar/gki979
A genomic approach to the identification and characterization of HOXA13 functional binding elements
Abstract
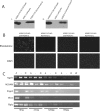
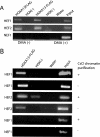
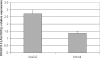
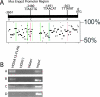
HOX proteins are important transcriptional regulators in mammalian embryonic development and are dysregulated in human cancers. However, there are few known direct HOX target genes and their mechanisms of regulation are incompletely understood. To isolate and characterize gene segments through which HOX proteins regulate transcription we used cesium chloride centrifugation-based chromatin purification and immunoprecipitation (ChIP). From NIH 3T3-derived HOXA13-FLAG expressing cells, 33% of randomly selected, ChIP clones were reproducibly enriched. Hox-enriched fragments (HEFs) were more AT-rich compared with cloned fragments that failed reproducible ChIP. All HEFs augmented transcription of a heterologous promoter upon coexpression with HOXA13. One HEF was from intron 2 of Enpp2, a gene highly upregulated in these cells and has been implicated in cell motility. Using Enpp2 as a candidate direct target, we identified three additional HEFs upstream of the transcription start site. HOXA13 upregulated transcription from an Enpp2 promoter construct containing these sites, and each site was necessary for full HOXA13-induced expression. Lastly, given that HOX proteins have been demonstrated to interact with histone deacetylases and/or CBP, we explored whether histone acetylation changed at Enpp2 upon HOXA13-induced activation. No change in the general histone acetylation state was observed. Our results support models in which occupation of multiple HOX binding sites is associated with highly activated genes.
Figures



Similar articles
-
Candidate downstream regulated genes of HOX group 13 transcription factors with and without monomeric DNA binding capability.Dev Biol. 2005 Mar 15;279(2):462-80. doi: 10.1016/j.ydbio.2004.12.015. Dev Biol. 2005. PMID: 15733672
-
Hoxd13 and Hoxa13 directly control the expression of the EphA7 Ephrin tyrosine kinase receptor in developing limbs.J Biol Chem. 2006 Jan 27;281(4):1992-9. doi: 10.1074/jbc.M510900200. Epub 2005 Nov 28. J Biol Chem. 2006. PMID: 16314414
-
Range of HOX/TALE superclass associations and protein domain requirements for HOXA13:MEIS interaction.Dev Biol. 2005 Jan 15;277(2):457-71. doi: 10.1016/j.ydbio.2004.10.004. Dev Biol. 2005. PMID: 15617687
-
Genomic approaches to understanding Hox gene function.Adv Genet. 2011;76:55-91. doi: 10.1016/B978-0-12-386481-9.00003-1. Adv Genet. 2011. PMID: 22099692 Review.
-
The homeodomain-containing proteins: an update on their interacting partners.Biochem Pharmacol. 1999 Dec 15;58(12):1851-7. doi: 10.1016/s0006-2952(99)00234-8. Biochem Pharmacol. 1999. PMID: 10591139 Review.
Cited by
-
E2F7 drives autotaxin/Enpp2 transcription via chromosome looping: Repression by p53 in murine but not in human carcinomas.FASEB J. 2023 Jul;37(7):e23058. doi: 10.1096/fj.202300838R. FASEB J. 2023. PMID: 37358838 Free PMC article.
-
Non-homeodomain regions of Hox proteins mediate activation versus repression of Six2 via a single enhancer site in vivo.Dev Biol. 2009 Nov 1;335(1):156-65. doi: 10.1016/j.ydbio.2009.08.020. Epub 2009 Aug 28. Dev Biol. 2009. PMID: 19716816 Free PMC article.
-
Loss of FOXF1 expression promotes human lung-resident mesenchymal stromal cell migration via ATX/LPA/LPA1 signaling axis.Sci Rep. 2020 Dec 4;10(1):21231. doi: 10.1038/s41598-020-77601-1. Sci Rep. 2020. PMID: 33277571 Free PMC article.
-
p100 Deficiency is insufficient for full activation of the alternative NF-κB pathway: TNF cooperates with p52-RelB in target gene transcription.PLoS One. 2012;7(8):e42741. doi: 10.1371/journal.pone.0042741. Epub 2012 Aug 6. PLoS One. 2012. PMID: 22880094 Free PMC article.
-
Recruitment of 5' Hoxa genes in the allantois is essential for proper extra-embryonic function in placental mammals.Development. 2012 Feb;139(4):731-9. doi: 10.1242/dev.075408. Epub 2012 Jan 4. Development. 2012. PMID: 22219351 Free PMC article.
References
-
- Veraksa A., Del Campo M., McGinnis W. Developmental patterning genes and their conserved functions: from model organisms to humans. Mol. Genet. Metab. 2000;69:85–100. - PubMed
-
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. - PubMed
-
- Grier D.G., Thompson A., Kwasniewska A., McGonigle G.J., Halliday H.L., Lappin T.R. The pathophysiology of HOX genes and their role in cancer. J. Pathol. 2005;205:154–171. - PubMed
-
- Biggin M.D., McGinnis W. Regulation of segmentation and segmental identity by Drosophila homeoproteins: the role of DNA binding in functional activity and specificity. Development. 1997;124:4425–4433. - PubMed
-
- Graba Y., Aragnol D., Pradel J. Drosophila Hox complex downstream targets and the function of homeotic genes. Bioessays. 1997;19:379–388. - PubMed

