Transcriptional intermediary factor 1alpha mediates physical interaction and functional synergy between the coactivator-associated arginine methyltransferase 1 and glucocorticoid receptor-interacting protein 1 nuclear receptor coactivators
- PMID: 16322096
- PMCID: PMC1626528
- DOI: 10.1210/me.2005-0393
Transcriptional intermediary factor 1alpha mediates physical interaction and functional synergy between the coactivator-associated arginine methyltransferase 1 and glucocorticoid receptor-interacting protein 1 nuclear receptor coactivators
Abstract
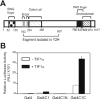
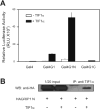
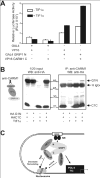
In previous studies transcriptional intermediary factor 1alpha (TIF1alpha) was identified as a direct binding partner and potential transcriptional coactivator for nuclear receptors (NRs) but its overexpression inhibited, rather than enhanced, transcriptional activation by NRs. Here we show that TIF1alpha bound to and enhanced the function of the C-terminal activation domain (AD) of coactivator associated arginine methyltransferase 1 (CARM1) and the N-terminal AD of glucocorticoid receptor-interacting protein 1 (GRIP1). Furthermore, although TIF1alpha had little or no NR coactivator activity by itself, it cooperated synergistically with GRIP1 and CARM1 to enhance NR-mediated transcription. Inhibition of endogenous TIF1alpha expression reduced transcriptional activation by the GRIP1 N-terminal domain but not by the CARM1 C-terminal domain, suggesting that TIF1alpha may be more important for mediating the activity of the former than the latter. Reduction of endogenous TIF1alpha levels also compromised the androgen-dependent induction of an endogenous target gene of the androgen receptor. Finally, TIF1alpha formed a ternary complex with the GRIP1 N-terminal and CARM1 C-terminal domains. Thus, we conclude that TIF1alpha cooperates with NR coactivators GRIP1 and CARM1 by forming a stable ternary complex with them and enhancing the AD function of one or both of them.
Conflict of interest statement
Disclosure of potential conflicts: C.T., C.-Y.O., K.K., R.L. have nothing to declare. M.R.S. receives royalties from Upstate Biotechnology, Inc. and received lecture fees from Bristol Myers-Squibb and Wyeth Pharmaceuticals.
Figures



References
-
- Beato M, Herrlich P, Schütz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. - PubMed
-
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. - PubMed
-
- McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. - PubMed
-
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. - PubMed
-
- Hermanson O, Glass CK, Rosenfeld MG. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol Metab. 2002;13:55–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

