Physical and functional interaction of DNA methyltransferase 3A with Mbd3 and Brg1 in mouse lymphosarcoma cells
- PMID: 16322236
- PMCID: PMC2241737
- DOI: 10.1158/0008-5472.CAN-05-1455
Physical and functional interaction of DNA methyltransferase 3A with Mbd3 and Brg1 in mouse lymphosarcoma cells
Retraction in
-
Retraction: Physical and Functional Interaction of DNA Methyltransferase 3A with Mbd3 and Brg1 in Mouse Lymphosarcoma Cells.Cancer Res. 2023 Dec 15;83(24):4182. doi: 10.1158/0008-5472.CAN-23-2344. Cancer Res. 2023. PMID: 38098452 Free PMC article. No abstract available.
Abstract
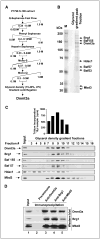
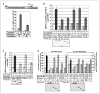
Dnmt3a and Dnmt3b are de novo DNA methyltransferases that also act as transcriptional repressors independent of methyltransferase activity. To elucidate the underlying mechanism of transcriptional repression, Dnmt3a was purified from mouse lymphosarcoma cells (P1798) by extensive fractionation on five different chromatographic matrices followed by glycerol density gradient centrifugation. Liquid chromatography electrospray tandem mass spectrometry analysis of Dnmt3a-associated polypeptides identified the methyl CpG binding protein Mbd3, histone deacetylase 1(Hdac1), and components of Brg1 complex (Brg1, Baf155, and Baf57) in the purified preparation. Association of Dnmt3a with Mbd3 and Brg1 was confirmed by coimmunoprecipitation and coimmunolocalization studies. Glutathione S-transferase pulldown assay showed that the NH2-terminal ATRX homology domain of Dnmt3a interacts with the methyl CpG binding domain of Mbd3 and with both bromo and ATPase domains of Brg1. Chromatin immunoprecipitation assay revealed that all three proteins are associated with transcriptionally silent methylated metallothionein (MT-I) promoter in the mouse lymphosarcoma cells. To understand the functional significance of their association with the promoter, their role on the MT-I promoter activity was analyzed by transient transfection assay. The results showed that Mbd3 and Dnmt3a specifically inhibited the methylated promoter, and the catalytic activity of Dnmt3a was dispensable for the suppression. In contrast, the wild-type but not the ATPase-inactive mutant of Brg1 suppressed MT-I promoter irrespective of its methylation status, implicating involvement of ATP-dependent chromatin remodeling in the process. Coexpression of two of the three interacting proteins at a time augmented their repressor function. This study shows physical and functional interaction of Dnmt3a with components of nucleosome remodeling machinery.
Figures




References
-
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyl-transferases. Nat Genet. 1998;19:219–20. - PubMed
-
- Okano M, Bell DW, Haber DA, Li E. DNA methyl-transferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. - PubMed
-
- Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–93. - PubMed
-
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. - PubMed
-
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

