Biochemical and genetic analysis of the yeast proteome with a movable ORF collection
- PMID: 16322557
- PMCID: PMC1315389
- DOI: 10.1101/gad.1362105
Biochemical and genetic analysis of the yeast proteome with a movable ORF collection
Abstract
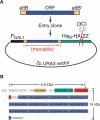
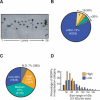
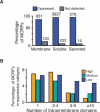
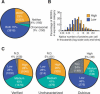
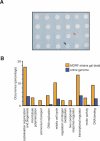
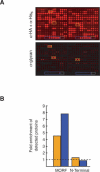
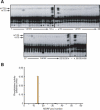
Functional analysis of the proteome is an essential part of genomic research. To facilitate different proteomic approaches, a MORF (moveable ORF) library of 5854 yeast expression plasmids was constructed, each expressing a sequence-verified ORF as a C-terminal ORF fusion protein, under regulated control. Analysis of 5573 MORFs demonstrates that nearly all verified ORFs are expressed, suggests the authenticity of 48 ORFs characterized as dubious, and implicates specific processes including cytoskeletal organization and transcriptional control in growth inhibition caused by overexpression. Global analysis of glycosylated proteins identifies 109 new confirmed N-linked and 345 candidate glycoproteins, nearly doubling the known yeast glycome.
Figures

References
-
- Akada, R., Yamamoto, J., and Yamashita, I. 1997. Screening and identification of yeast sequences that cause growth inhibition when overexpressed. Mol. Gen. Genet. 254: 267-274. - PubMed
-
- Apweiler, R., Hermjakob, H., and Sharon, N. 1999. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473: 4-8. - PubMed
-
- Bader, G.D., Heilbut, A., Andrews, B., Tyers, M., Hughes, T., and Boone, C. 2003. Functional genomics and proteomics: Charting a multidimensional map of the yeast cell. Trends Cell. Biol. 13: 344-356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
