Loss of function of def selectively up-regulates Delta113p53 expression to arrest expansion growth of digestive organs in zebrafish
- PMID: 16322560
- PMCID: PMC1315396
- DOI: 10.1101/gad.1366405
Loss of function of def selectively up-regulates Delta113p53 expression to arrest expansion growth of digestive organs in zebrafish
Abstract
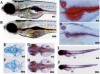
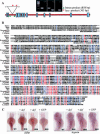
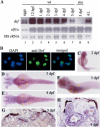
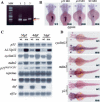
Transcription factor p53 forms a network with associated factors to regulate the cell cycle and apoptosis in response to environmental stresses. However, there is currently no direct genetic evidence to show if or how the p53 pathway functions during organogenesis. Here we present evidence to show that the zebrafish def (digestive-organ expansion factor) gene encodes a novel pan-endoderm-specific factor. A loss-of-function mutation in def confers hypoplastic digestive organs and selectively up-regulates the expression of Delta113p53, counterpart to a newly identified isoform of p53 produced by an alternative internal promoter in intron 4 of the p53 gene in human. The increased Delta113p53 expression is limited to within the mutant digestive organs, and this increase selectively induces the expression of p53-responsive genes to trigger the arrest of the cell cycle but not apoptosis, resulting in compromised organ growth in the mutant. Our data demonstrate that, while induction of expression of p53 and/or its isoforms is crucial to suppress abnormal cell growth, Delta113p53 is tightly regulated by an organ/tissue-specific factor Def, especially during organogenesis, to prevent adverse inhibition of organ/tissue growth.
Figures


References
-
- Armstrong, J.F., Kaufman, M.H., Harrison, D.J., and Clarke, A.R. 1995. High-frequency developmental abnormalities in p53-deficient mice. Curr. Biol. 5: 931-936. - PubMed
-
- Ball, K.L. 1997. p21: Structure and functions associated with cyclin-CDK binding. Prog. Cell Cycle Res. 3: 125-134. - PubMed
-
- Benard, J., Douc-Rasy, S., and Ahomadegbe, J.C. 2003. TP53 family members and human cancers. Hum. Mutat. 21: 182-191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
