Identification of surface-exposed components of MOMP of Chlamydia trachomatis serovar F
- PMID: 16322562
- PMCID: PMC2242375
- DOI: 10.1110/ps.051616206
Identification of surface-exposed components of MOMP of Chlamydia trachomatis serovar F
Abstract
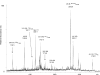
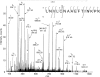
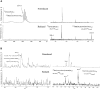
The identification of surface-exposed components of the major outer membrane protein (MOMP) of Chlamydia is critical for modeling its three-dimensional structure, as well as for understanding the role of MOMP in the pathogenesis of Chlamydia-related diseases. MOMP contains four variable domains (VDs). In this study, VDII and VDIV of Chlamydia trachomatis serovar F were proven to be surface-located by immuno-dot blot assay using monoclonal antibodies (MAbs). Two proteases, trypsin and endoproteinase Glu-C, were applied to digest the intact elementary body of serovar F under native conditions to reveal the surface-located amino acids. The resulting peptides were separated by SDS-PAGE and probed with MAbs against these VDs. N-terminal amino acid sequencing revealed: (1) The Glu-C cleavage sites were located within VDI (at Glu61) and VDIII (at Glu225); (2) the trypsin cleavage sites were found at Lys79 in VDI and at Lys224 in VDIII. The tryptic peptides were then isolated by HPLC and analyzed with a matrix-assisted laser desorption/ionization time-of-flight mass spectrometer and a quadrupole-orthogonal-TOF mass spectrometer coupled with a capillary liquid chromatograph. Masses and fragmentation patterns that correlated to the peptides cleaved from VDI and VDIII regions, and C-terminal peptides Ser333-Arg358 and Ser333-Lys350 were observed. This result demonstrated that these regions are surface-exposed. Data derived from comparison of nonreduced outer membrane complex proteolytic fragments with their reduced fractions revealed that Cys26, 29, 33, 116, 208, and 337 were involved in disulfide bonds, and Cys26 and 337, and 116 and 208 were paired. Based on these data, a new two-dimensional model is proposed.
Figures



Similar articles
-
Prediction of the membrane-spanning beta-strands of the major outer membrane protein of Chlamydia.Protein Sci. 2002 Jul;11(7):1854-61. doi: 10.1110/ps.3650102. Protein Sci. 2002. PMID: 12070338 Free PMC article.
-
Characterization of the disulfide bonds and free cysteine residues of the Chlamydia trachomatis mouse pneumonitis major outer membrane protein.Biochemistry. 2005 Apr 26;44(16):6250-6. doi: 10.1021/bi047775v. Biochemistry. 2005. PMID: 15835913
-
Structural and functional analyses of the major outer membrane protein of Chlamydia trachomatis.J Bacteriol. 2007 Sep;189(17):6222-35. doi: 10.1128/JB.00552-07. Epub 2007 Jun 29. J Bacteriol. 2007. PMID: 17601785 Free PMC article.
-
Determination of neutralizing epitopes in variable domains I and IV of the major outer-membrane protein from Chlamydia trachomatis serovar K.Microbiology (Reading). 1994 Sep;140 ( Pt 9):2481-7. doi: 10.1099/13500872-140-9-2481. Microbiology (Reading). 1994. PMID: 7524957
-
Differential effect of trypsin on infectivity of Chlamydia trachomatis: loss of infectivity requires cleavage of major outer membrane protein variable domains II and IV.Infect Immun. 1988 Aug;56(8):2094-100. doi: 10.1128/iai.56.8.2094-2100.1988. Infect Immun. 1988. PMID: 2456271 Free PMC article.
Cited by
-
The early gene product EUO is a transcriptional repressor that selectively regulates promoters of Chlamydia late genes.Mol Microbiol. 2012 Jun;84(6):1097-107. doi: 10.1111/j.1365-2958.2012.08077.x. Epub 2012 May 25. Mol Microbiol. 2012. PMID: 22624851 Free PMC article.
-
Diversity of σ66-Specific Promoters Contributes to Regulation of Developmental Gene Expression in Chlamydia trachomatis.J Bacteriol. 2023 Jan 26;205(1):e0031022. doi: 10.1128/jb.00310-22. Epub 2023 Jan 4. J Bacteriol. 2023. PMID: 36598485 Free PMC article.
-
Spectroscopic analysis of chlamydial major outer membrane protein in support of structure elucidation.Protein Sci. 2018 Nov;27(11):1923-1941. doi: 10.1002/pro.3501. Protein Sci. 2018. PMID: 30144190 Free PMC article.
-
Chlamydia trachomatis: when the virulence-associated genome backbone imports a prevalence-associated major antigen signature.Microb Genom. 2019 Nov;5(11):e000313. doi: 10.1099/mgen.0.000313. Microb Genom. 2019. PMID: 31697227 Free PMC article.
-
Overexpression of the Bam Complex Improves the Production of Chlamydia trachomatis MOMP in the E. coli Outer Membrane.Int J Mol Sci. 2022 Jul 2;23(13):7393. doi: 10.3390/ijms23137393. Int J Mol Sci. 2022. PMID: 35806397 Free PMC article.
References
-
- Aitken, A. and Learmonth, M. 1996. Carboxymethylation of cysteine using iodoacetamide/iodoacetic acid. In The protein protocols handbook (ed. J.M. Walker), pp. 339–340. Humana Press, Totawa, NJ.
-
- Baumann, M. and Meri, S. 2004. Techniques for studying protein heterogeneity and post-translational modifications. Expert Rev. Proteomics 1: 207–217. - PubMed
-
- Boulain, J.C., Charbit, A., and Hofnung, M. 1986. Mutagenesis by random linker insertion into the lamB gene of Escherichia coli K12. Mol. Gen. 205: 339–348. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

