Simulation of pH-dependent edge strand rearrangement in human beta-2 microglobulin
- PMID: 16322574
- PMCID: PMC2242376
- DOI: 10.1110/ps.051814306
Simulation of pH-dependent edge strand rearrangement in human beta-2 microglobulin
Abstract
Amyloid fibrils formed from unrelated proteins often share morphological similarities, suggesting common biophysical mechanisms for amyloidogenesis. Biochemical studies of human beta-2 microglobulin (beta2M) have shown that its transition from a water-soluble protein to insoluble aggregates can be triggered by low pH. Additionally, biophysical measurements of beta2M using NMR have identified residues of the protein that participate in the formation of amyloid fibrils. The crystal structure of monomeric human beta2M determined at pH 5.7 shows that one of its edge beta-strands (strand D) adopts a conformation that differs from other structures of the same protein obtained at higher pH. This alternate beta-strand arrangement lacks a beta-bulge, which may facilitate protein aggregation through intermolecular beta-sheet association. To explore whether the pH change may yield the observed conformational difference, molecular dynamics simulations of beta2M were performed. The effects of pH were modeled by specifying the protonation states of Asp, Glu, and His, as well as the C terminus of the main chain. The bulged conformation of strand D is preferred at medium pH (pH 5-7), whereas at low pH (pH < 4) the straight conformation is observed. Therefore, low pH may stabilize the straight conformation of edge strand D and thus increase the amyloidogenicity of beta2M.
Figures




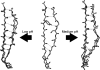
Similar articles
-
D-strand perturbation and amyloid propensity in beta-2 microglobulin.FEBS J. 2011 Jul;278(13):2349-58. doi: 10.1111/j.1742-4658.2011.08157.x. Epub 2011 May 31. FEBS J. 2011. PMID: 21569201
-
A recurrent D-strand association interface is observed in β-2 microglobulin oligomers.FEBS J. 2012 Mar;279(6):1131-43. doi: 10.1111/j.1742-4658.2012.08510.x. Epub 2012 Feb 23. FEBS J. 2012. PMID: 22289140
-
A single disulfide bond differentiates aggregation pathways of beta2-microglobulin.J Mol Biol. 2005 Nov 25;354(2):473-82. doi: 10.1016/j.jmb.2005.09.075. Epub 2005 Oct 7. J Mol Biol. 2005. PMID: 16242719
-
Molecular interactions in the formation and deposition of beta2-microglobulin-related amyloid fibrils.Amyloid. 2005 Mar;12(1):15-25. doi: 10.1080/13506120500032352. Amyloid. 2005. PMID: 16076607 Review.
-
Solution structure of beta(2)-microglobulin and insights into fibrillogenesis.Biochim Biophys Acta. 2005 Nov 10;1753(1):76-84. doi: 10.1016/j.bbapap.2005.07.003. Epub 2005 Jul 21. Biochim Biophys Acta. 2005. PMID: 16081329 Review.
Cited by
-
The second Ca2+-binding domain of the Na+ Ca2+ exchanger is essential for regulation: crystal structures and mutational analysis.Proc Natl Acad Sci U S A. 2007 Nov 20;104(47):18467-72. doi: 10.1073/pnas.0707417104. Epub 2007 Oct 25. Proc Natl Acad Sci U S A. 2007. PMID: 17962412 Free PMC article.
-
Insights into the role of the beta-2 microglobulin D-strand in amyloid propensity revealed by mass spectrometry.Mol Biosyst. 2014 Mar 4;10(3):412-20. doi: 10.1039/c3mb70420c. Epub 2013 Dec 12. Mol Biosyst. 2014. PMID: 24336936 Free PMC article.
References
-
- Arora, A., Ha, C., and Park, C.B. 2004a. Inhibition of insulin amyloid formation by small stress molecules. FEBS Lett. 564: 121–125. - PubMed
-
- Baptista, A.M., Martel, P.J., and Petersen, S.B. 1997. Simulation of protein conformational freedom as a function of pH: Constant-pH molecular dynamics using implicit titration. Proteins 27: 523–544. - PubMed
-
- Baptista, A.M., Teixeira, V.H., and Soares, C.M. 2002. Constant-pH molecular dynamics using stochastic titration. J. Chem. Phys. 117: 4184–4200.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

