Sulf-2, a proangiogenic heparan sulfate endosulfatase, is upregulated in breast cancer
- PMID: 16331886
- PMCID: PMC1502017
- DOI: 10.1593/neo.05496
Sulf-2, a proangiogenic heparan sulfate endosulfatase, is upregulated in breast cancer
Abstract
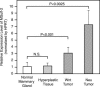
Sulf-2 is an endosulfatase with activity against glucosamine-6-sulfate modifications within subregions of intact heparin. The enzyme has the potential to modify the sulfation status of extracellular heparan sulfate proteoglycan (HSPG) glycosaminoglycan chains and thereby to regulate interactions with HSPG-binding proteins. In the present investigation, data mining from published studies was employed to establish Sulf-2 mRNA upregulation in human breast cancer. We further found that cultured breast carcinoma cells expressed Sulf-2 mRNA and released enzymatically active proteins into conditioned medium. In two mouse models of mammary carcinoma, Sulf-2 mRNA was upregulated in comparison to its expression in normal mammary gland. Although mRNA was present in normal tissues, Sulf-2 protein was undetectable; it was, however, detected in some premalignant lesions and in tumors. The protein was localized to the epithelial cells of the tumors. In support of the possible mechanistic relevance of Sulf-2 upregulation in tumors, purified recombinant Sulf-2 promoted angiogenesis in the chick chorioallantoic membrane assay.
Figures







References
-
- Sasisekharan R, Shriver Z, Venkataraman G, Narayanasami U. Roles of heparan-sulphate glycosaminoglycans in cancer. Nat Rev Cancer. 2002;2:521–528. - PubMed
-
- Esko JD, Selleck SB. ORDER OUT OF CHAOS: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. - PubMed
-
- Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, Mohammadi M. Crystal structure of a ternary FGF–FGFR–heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–750. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
