Mechanistic characterization of the MSDH (methylmalonate semialdehyde dehydrogenase) from Bacillus subtilis
- PMID: 16332250
- PMCID: PMC1409689
- DOI: 10.1042/BJ20051525
Mechanistic characterization of the MSDH (methylmalonate semialdehyde dehydrogenase) from Bacillus subtilis
Abstract
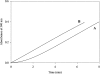
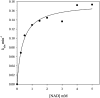
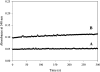
Homotetrameric MSDH (methylmalonate semialdehyde dehydrogenase) from Bacillus subtilis catalyses the NAD-dependent oxidation of MMSA (methylmalonate semialdehyde) and MSA (malonate semialdehyde) into PPCoA (propionyl-CoA) and acetyl-CoA respectively via a two-step mechanism. In the present study, a detailed mechanistic characterization of the MSDH-catalysed reaction has been carried out. The results suggest that NAD binding elicits a structural imprinting of the apoenzyme, which explains the marked lag-phase observed in the activity assay. The enzyme also exhibits a half-of-the-sites reactivity, with two subunits being active per tetramer. This result correlates well with the presence of two populations of catalytic Cys302 in both the apo- and holo-enzymes. Binding of NAD causes a decrease in reactivity of the two Cys302 residues belonging to the two active subunits and a pKapp shift from approx. 8.8 to 8.0. A study of the rate of acylation as a function of pH revealed a decrease in the pKapp of the two active Cys302 residues to approx. 5.5. Taken to-gether, these results support a sequential Cys302 activation process with a pKapp shift from approx. 8.8 in the apo-form to 8.0 in the binary complex and finally to approx. 5.5 in the ternary complex. The rate-limiting step is associated with the b-decarboxylation process which occurs on the thioacylenzyme intermediate after NADH release and before transthioesterification. These data also indicate that bicarbonate, the formation of which is enzyme-catalysed, is the end-product of the reaction.
Figures






References
-
- Liu Z. J., Sun Y. J., Rose J., Chung Y. J., Hsiao C. D., Chang W. R., Kuo I., Perozich J., Lindahl R., Hempel J., Wang B. C. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat. Struct. Biol. 1997;4:317–326. - PubMed
-
- Steinmetz C. G., Xie P., Weiner H., Hurley T. D. Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure. 1997;5:701–711. - PubMed
-
- Cobessi D., Tete-Favier F., Marchal S., Azza S., Branlant G., Aubry A. Apo and holo crystal structures of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans. J. Mol. Biol. 1999;290:161–173. - PubMed
-
- Farres J., Wang T. T., Cunningham S. J., Weiner H. Investigation of the active site cysteine residue of rat liver mitochondrial aldehyde dehydrogenase by site-directed mutagenesis. Biochemistry. 1995;34:2592–2598. - PubMed
-
- Vedadi M., Szittner R., Smillie L., Meighen E. Involvement of cysteine 289 in the catalytic activity of an NADP+-specific fatty aldehyde dehydrogenase from Vibrio harveyi. Biochemistry. 1995;34:16725–16732. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

