Cloning and characterization of two xyloglucanases from Paenibacillus sp. strain KM21
- PMID: 16332739
- PMCID: PMC1317386
- DOI: 10.1128/AEM.71.12.7670-7678.2005
Cloning and characterization of two xyloglucanases from Paenibacillus sp. strain KM21
Abstract
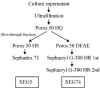
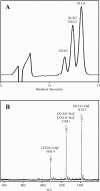
Two xyloglucan-specific endo-beta-1,4-glucanases (xyloglucanases [XEGs]), XEG5 and XEG74, with molecular masses of 40 kDa and 105 kDa, respectively, were isolated from the gram-positive bacterium Paenibacillus sp. strain KM21, which degrades tamarind seed xyloglucan. The genes encoding these XEGs were cloned and sequenced. Based on their amino acid sequences, the catalytic domains of XEG5 and XEG74 were classified in the glycoside hydrolase families 5 and 74, respectively. XEG5 is the first xyloglucanase belonging to glycoside hydrolase family 5. XEG5 lacks a carbohydrate-binding module, while XEG74 has an X2 module and a family 3 type carbohydrate-binding module at its C terminus. The two XEGs were expressed in Escherichia coli, and recombinant forms of the enzymes were purified and characterized. Both XEGs had endoglucanase active only toward xyloglucan and not toward Avicel, carboxymethylcellulose, barley beta-1,3/1,4-glucan, or xylan. XEG5 is a typical endo-type enzyme that randomly cleaves the xyloglucan main chain, while XEG74 has dual endo- and exo-mode activities or processive endo-mode activity. XEG5 digested the xyloglucan oligosaccharide XXXGXXXG to produce XXXG, whereas XEG74 digestion of XXXGXXXG resulted in XXX, XXXG, and GXXXG, suggesting that this enzyme cleaves the glycosidic bond of unbranched Glc residues. Analyses using various oligosaccharide structures revealed that unique structures of xyloglucan oligosaccharides can be prepared with XEG74.
Figures






References
-
- Blanco, A., P. Díaz, J. Martínez, T. Vidal, A. L. Torres, and F. I. J. Pastor. 1998. Cloning of a new endoglucanase gene from Bacillus sp. BP-23 and characterisation of the enzyme. Performance in paper manufacture from cereal straw. Appl. Microbiol. Biotechnol. 50:48-54. - PubMed
-
- Reference deleted.
-
- Fry, S. C. 1989. The structure and functions of xyloglucan. J. Exp. Bot. 40:1-11.
-
- Fry, S. C., W. S. York, P. Albersheim, A. Darvill, T. Hayashi, J. P. Joseleau, Y. Kato, E. P. Lorences, G. A. Maclachlan, M. McNeil, A. J. Mort, J. S. G. Reid, H. U. Seitz, R. R. Selvendran, A. G. J. Voragen, and A. R. White. 1993. An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol. Plant. 89:1-3.
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

