At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies
- PMID: 16332745
- PMCID: PMC1317345
- DOI: 10.1128/AEM.71.12.7724-7736.2005
At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies
Abstract
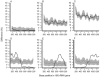
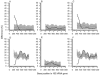
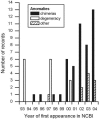
A new method for detecting chimeras and other anomalies within 16S rRNA sequence records is presented. Using this method, we screened 1,399 sequences from 19 phyla, as defined by the Ribosomal Database Project, release 9, update 22, and found 5.0% to harbor substantial errors. Of these, 64.3% were obvious chimeras, 14.3% were unidentified sequencing errors, and 21.4% were highly degenerate. In all, 11 phyla contained obvious chimeras, accounting for 0.8 to 11% of the records for these phyla. Many chimeras (43.1%) were formed from parental sequences belonging to different phyla. While most comprised two fragments, 13.7% were composed of at least three fragments, often from three different sources. A separate analysis of the Bacteroidetes phylum (2,739 sequences) also revealed 5.8% records to be anomalous, of which 65.4% were apparently chimeric. Overall, we conclude that, as a conservative estimate, 1 in every 20 public database records is likely to be corrupt. Our results support concerns recently expressed over the quality of the public repositories. With 16S rRNA sequence data increasingly playing a dominant role in bacterial systematics and environmental biodiversity studies, it is vital that steps be taken to improve screening of sequences prior to submission. To this end, we have implemented our method as a program with a simple-to-use graphic user interface that is capable of running on a range of computer platforms. The program is called Pintail, is released under the terms of the GNU General Public License open source license, and is freely available from our website at http://www.cardiff.ac.uk/biosi/research/biosoft/.
Figures



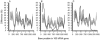

References
-
- Cole, J., B. Chai, T. Marsh, R. Farris, Q. Wang, S. Kulum, S. Chandra, D. McGarrell, T. Schmidt, G. Garrity, and J. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. - PMC - PubMed
-
- Fox, J. L. 2005. Ribosomal gene milestone met, already left in dust. ASM News 71:6-7.
-
- Garrity, G. M., M. Winters, A. W. Kuo, and D. Searles. 2002. Taxonomic outline of the prokaryotes, p. 49-66. Bergey's manual of systematic bacteriology, 2nd ed. Springer-Verlag, New York, N.Y.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

