A broad-host-range, generalized transducing phage (SN-T) acquires 16S rRNA genes from different genera of bacteria
- PMID: 16332816
- PMCID: PMC1317401
- DOI: 10.1128/AEM.71.12.8301-8304.2005
A broad-host-range, generalized transducing phage (SN-T) acquires 16S rRNA genes from different genera of bacteria
Abstract
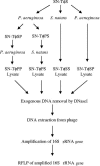
Genomic analysis has revealed heterogeneity among bacterial 16S rRNA gene sequences within a single species; yet the cause(s) remains uncertain. Generalized transducing bacteriophages have recently gained recognition for their abundance as well as their ability to affect lateral gene transfer and to harbor bacterial 16S rRNA gene sequences. Here, we demonstrate the ability of broad-host-range, generalized transducing phages to acquire 16S rRNA genes and gene sequences. Using PCR and primers specific to conserved regions of the 16S rRNA gene, we have found that generalized transducing phages (D3112, UT1, and SN-T), but not specialized transducing phages (D3), acquired entire bacterial 16S rRNA genes. Furthermore, we show that the broad-host-range, generalized transducing phage SN-T is capable of acquiring the 16S rRNA gene from two different genera: Sphaerotilus natans, the host from which SN-T was originally isolated, and Pseudomonas aeruginosa. In sequential infections, SN-T harbored only 16S rRNA gene sequences of the final host as determined by restriction fragment length polymorphism analysis. The frequency of 16S rRNA gene sequences in SN-T populations was determined to be 1 x 10(-9) transductants/PFU. Our findings further implicate transduction in the horizontal transfer of 16S rRNA genes between different species or genera of bacteria.
Figures

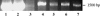
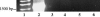


Similar articles
-
Method for host-independent detection of generalized transducing bacteriophages in natural habitats.Appl Environ Microbiol. 2001 Apr;67(4):1490-3. doi: 10.1128/AEM.67.4.1490-1493.2001. Appl Environ Microbiol. 2001. PMID: 11282595 Free PMC article.
-
Transduction of a freshwater microbial community by a new Pseudomonas aeruginosa generalized transducing phage, UT1.Mol Ecol. 1994 Apr;3(2):121-6. doi: 10.1111/j.1365-294x.1994.tb00112.x. Mol Ecol. 1994. PMID: 8019688
-
Analysis of transduction in wastewater bacterial populations by targeting the phage-derived 16S rRNA gene sequences.FEMS Microbiol Ecol. 2011 Apr;76(1):100-8. doi: 10.1111/j.1574-6941.2010.01034.x. Epub 2011 Jan 19. FEMS Microbiol Ecol. 2011. PMID: 21223328
-
[Role of horizontal gene transfer by bacteriophages in the origin of pathogenic bacteria].Genetika. 2003 May;39(5):595-620. Genetika. 2003. PMID: 12838609 Review. Russian.
-
Bacteriophages and genetic mobilization in sewage and faecally polluted environments.Microb Biotechnol. 2011 Nov;4(6):725-34. doi: 10.1111/j.1751-7915.2011.00264.x. Epub 2011 Apr 27. Microb Biotechnol. 2011. PMID: 21535427 Free PMC article. Review.
Cited by
-
Revisiting bacterial phylogeny: Natural and experimental evidence for horizontal gene transfer of 16S rRNA.Mob Genet Elements. 2013 Jan 1;3(1):e24210. doi: 10.4161/mge.24210. Mob Genet Elements. 2013. PMID: 23734299 Free PMC article.
-
Role of Bacterial-Fungal Consortium for Enhancement in the Degradation of Industrial Dyes.Curr Genomics. 2020 May;21(4):283-294. doi: 10.2174/1389202921999200505082901. Curr Genomics. 2020. PMID: 33071621 Free PMC article.
-
Reservoir of bacterial exotoxin genes in the environment.Int J Microbiol. 2010;2010:754368. doi: 10.1155/2010/754368. Epub 2011 Jan 9. Int J Microbiol. 2010. PMID: 21318166 Free PMC article.
-
Virus-host interactions predictor (VHIP): Machine learning approach to resolve microbial virus-host interaction networks.PLoS Comput Biol. 2024 Sep 18;20(9):e1011649. doi: 10.1371/journal.pcbi.1011649. eCollection 2024 Sep. PLoS Comput Biol. 2024. PMID: 39292721 Free PMC article.
-
Insights and inferences about integron evolution from genomic data.BMC Genomics. 2008 May 31;9:261. doi: 10.1186/1471-2164-9-261. BMC Genomics. 2008. PMID: 18513439 Free PMC article.
References
-
- Clayton, R. A., G. Sutton, P. S. Hinkle, Jr., C. Bult, and C. Field. 1995. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int. J. Syst. Bacteriol. 45:595-599. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

