Characterization of microbial communities found in the human vagina by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes
- PMID: 16332868
- PMCID: PMC1317315
- DOI: 10.1128/AEM.71.12.8729-8737.2005
Characterization of microbial communities found in the human vagina by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes
Abstract
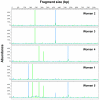
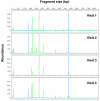
To define and monitor the structure of microbial communities found in the human vagina, a cultivation-independent approach based on analyses of terminal restriction fragment length polymorphisms (T-RFLP) of 16S rRNA genes was developed and validated. Sixteen bacterial strains commonly found in the human vagina were used to construct model communities that were subsequently used to develop efficient means for the isolation of genomic DNA and an optimal strategy for T-RFLP analyses. The various genera in the model community could best be resolved by digesting amplicons made using bacterial primers 8f and 926r with HaeIII; fewer strains could be resolved using other primer-enzyme combinations, and no combination successfully distinguished certain species of the same genus. To demonstrate the utility of the approach, samples from five women that had been collected over a 2-month period were analyzed. Differences and similarities among the vaginal microbial communities of the women were readily apparent. The T-RFLP data suggest that the communities of three women were dominated by a single phylotype, most likely species of Lactobacillus. In contrast, the communities of two other women included numerically abundant populations that differed from Lactobacillus strains whose 16S rRNA genes had been previously determined. The T-RFLP profiles of samples from all the women were largely invariant over time, indicating that the kinds and abundances of the numerically dominant populations were relatively stable throughout two menstrual cycles. These findings show that T-RFLP of 16S rRNA genes can be used to compare vaginal microbial communities and gain information about the numerically dominant populations that are present.
Figures


References
-
- Antonio, M. A., S. E. Hawes, and S. L. Hillier. 1999. The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species. J. Infect. Dis. 180:1950-1956. - PubMed
-
- Bartlett, J. G., A. B. Onderdonk, E. Drude, C. Goldstein, M. Anderka, S. Alpert, and W. M. McCormack. 1977. Quantitative bacteriology of the vaginal flora. J. Infect. Dis. 136:271-277. - PubMed
-
- Bartlett, J. G., and B. F. Polk. 1984. Bacterial flora of the vagina: quantitative study. Rev. Infect. Dis. Suppl. 6:S67-S72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

