Differential protein-DNA interactions at the promoter and enhancer regions of developmentally regulated U4 snRNA genes
- PMID: 1633438
- PMCID: PMC6057390
Differential protein-DNA interactions at the promoter and enhancer regions of developmentally regulated U4 snRNA genes
Abstract
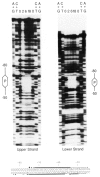
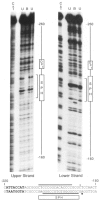
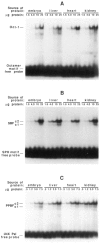
In the chicken genome there are two closely-linked genes, U4B and U4X, that code for different sequence variants of U4 small nuclear RNA (snRNA). Both genes are expressed with nearly equal efficiency in the early embryo, but U4X gene expression is specifically down-regulated relative to U4B as development proceeds. At the present time, little is known about the mechanisms that regulate differential expression of snRNA genes. We have now identified a novel chicken factor, PPBF, that binds sequence-specifically in vitro to the proximal regulatory region of the U4X gene, but not to the proximal region of the U4B gene. PPBF is itself regulated during development and may therefore be a key factor involved in differentially regulating U4X gene transcription relative to U4B. The U4X and U4B enhancers contain distinct sequence variants of two essential motifs (octamer and SPH). The Oct-1 transcription factor binds with similar affinities to both the U4X and U4B octamer motifs. However, a second essential snRNA enhancer-binding protein, SBF, has a 20- to 30-fold lower affinity for the SPH motif in the U4X enhancer than for the homologous SPH motif in the U4B enhancer. A potential role therefore exists for SBF, as well as PPBF, in the preferential down-regulation of the U4X RNA gene during chicken development.
Figures






References
-
- Dahlberg J. E. and Lund E. (1988), in Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles (Birnstiel M. L., ed.), Springer Verlag KG, Heidelberg, pp. 38–70.
-
- Forbes D. J., Kirschner M. W., Caput D., Dahlberg J. E., and Lund E. (1984), Cell 38, 681–689. - PubMed
-
- Gunderson S. I., Knuth M. W., and Burgess R. R. (1990), Genes Dev 4, 2048–2060. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
