Identification of three mammalian proteins that bind to the yeast TATA box protein TFIID
- PMID: 1633441
- PMCID: PMC6057387
Identification of three mammalian proteins that bind to the yeast TATA box protein TFIID
Abstract
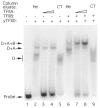
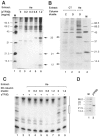
The TATA box binding transcription factor TFIID of S. cerevisiae was used as a ligand for affinity chromatography. Polypeptides that bind specifically to yeast TFIID (TFIID-associated proteins, DAPs) were purified from human HeLa (heDAPs) and calf thymus (ctDAPs) whole cell extracts. Both heDAP and ctDAP fractions altered the binding of TFIID to the TATA element, and substituted for the TFIIA transcription activity in a reconstituted in vitro system. The heDAP fraction also behaved like TFIIA in its ability to form a promoter-TFIID-TFIIA complex and to recruit TFIIB to such a complex. The interaction of DAPs with TFIID can confer heat-resistance (47 degrees C) on recombinant yeast or human TFIID. SDS-PAGE analysis revealed that three polypeptides from HeLa extracts specifically bound to yTFIID columns (heDAP35, heDAP21, and heDAP12). These data suggest that a multi-subunit transcription factor with the properties of TFIIA can bind to TFIID in the absence of DNA.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
