Conserved and variable functions of the sigmaE stress response in related genomes
- PMID: 16336047
- PMCID: PMC1312014
- DOI: 10.1371/journal.pbio.0040002
Conserved and variable functions of the sigmaE stress response in related genomes
Abstract
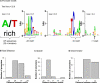
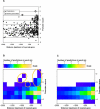
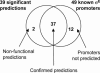
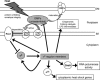
Bacteria often cope with environmental stress by inducing alternative sigma (sigma) factors, which direct RNA polymerase to specific promoters, thereby inducing a set of genes called a regulon to combat the stress. To understand the conserved and organism-specific functions of each sigma, it is necessary to be able to predict their promoters, so that their regulons can be followed across species. However, the variability of promoter sequences and motif spacing makes their prediction difficult. We developed and validated an accurate promoter prediction model for Escherichia coli sigmaE, which enabled us to predict a total of 89 unique sigmaE-controlled transcription units in E. coli K-12 and eight related genomes. SigmaE controls the envelope stress response in E. coli K-12. The portion of the regulon conserved across genomes is functionally coherent, ensuring the synthesis, assembly, and homeostasis of lipopolysaccharide and outer membrane porins, the key constituents of the outer membrane of Gram-negative bacteria. The larger variable portion is predicted to perform pathogenesis-associated functions, suggesting that sigmaE provides organism-specific functions necessary for optimal host interaction. The success of our promoter prediction model for sigmaE suggests that it will be applicable for the prediction of promoter elements for many alternative sigma factors.
Figures

Comment in
-
Bacterial stress responses: what doesn't kill them can make then stronger.PLoS Biol. 2006 Jan;4(1):e23. doi: 10.1371/journal.pbio.0040023. PLoS Biol. 2006. PMID: 16535775 Free PMC article.
References
-
- Burden S, Lin YX, Zhang R. Improving promoter prediction for the NNPP2.2 algorithm: A case study using Escherichia coli DNA sequences. Bioinformatics. 2004;1:601–607. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

