Common themes and cell type specific variations of higher order chromatin arrangements in the mouse
- PMID: 16336643
- PMCID: PMC1325247
- DOI: 10.1186/1471-2121-6-44
Common themes and cell type specific variations of higher order chromatin arrangements in the mouse
Abstract
Background: Similarities as well as differences in higher order chromatin arrangements of human cell types were previously reported. For an evolutionary comparison, we now studied the arrangements of chromosome territories and centromere regions in six mouse cell types (lymphocytes, embryonic stem cells, macrophages, fibroblasts, myoblasts and myotubes) with fluorescence in situ hybridization and confocal laser scanning microscopy. Both species evolved pronounced differences in karyotypes after their last common ancestors lived about 87 million years ago and thus seem particularly suited to elucidate common and cell type specific themes of higher order chromatin arrangements in mammals.
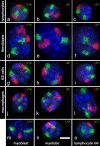
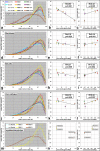
Results: All mouse cell types showed non-random correlations of radial chromosome territory positions with gene density as well as with chromosome size. The distribution of chromosome territories and pericentromeric heterochromatin changed during differentiation, leading to distinct cell type specific distribution patterns. We exclude a strict dependence of these differences on nuclear shape. Positional differences in mouse cell nuclei were less pronounced compared to human cell nuclei in agreement with smaller differences in chromosome size and gene density. Notably, the position of chromosome territories relative to each other was very variable.
Conclusion: Chromosome territory arrangements according to chromosome size and gene density provide common, evolutionary conserved themes in both, human and mouse cell types. Our findings are incompatible with a previously reported model of parental genome separation.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

