Uneven distribution of expressed sequence tag loci on maize pachytene chromosomes
- PMID: 16339046
- PMCID: PMC1356135
- DOI: 10.1101/gr.4249906
Uneven distribution of expressed sequence tag loci on maize pachytene chromosomes
Abstract
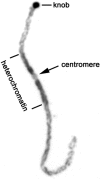
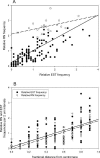
Examining the relationships among DNA sequence, meiotic recombination, and chromosome structure at a genome-wide scale has been difficult because only a few markers connect genetic linkage maps with physical maps. Here, we have positioned 1195 genetically mapped expressed sequence tag (EST) markers onto the 10 pachytene chromosomes of maize by using a newly developed resource, the RN-cM map. The RN-cM map charts the distribution of crossing over in the form of recombination nodules (RNs) along synaptonemal complexes (SCs, pachytene chromosomes) and allows genetic cM distances to be converted into physical micrometer distances on chromosomes. When this conversion is made, most of the EST markers used in the study are located distally on the chromosomes in euchromatin. ESTs are significantly clustered on chromosomes, even when only euchromatic chromosomal segments are considered. Gene density and recombination rate (as measured by EST and RN frequencies, respectively) are strongly correlated. However, crossover frequencies for telomeric intervals are much higher than was expected from their EST frequencies. For pachytene chromosomes, EST density is about fourfold higher in euchromatin compared with heterochromatin, while DNA density is 1.4 times higher in heterochromatin than in euchromatin. Based on DNA density values and the fraction of pachytene chromosome length that is euchromatic, we estimate that approximately 1500 Mbp of the maize genome is in euchromatin. This overview of the organization of the maize genome will be useful in examining genome and chromosome evolution in plants.
Figures

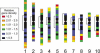
References
-
- Akhunov, E.D., Akhunova, A.R., Linkiewicz, A.M., Dubcovsky, J., Hummel, D., Lazo, G.R., Chao, S., Anderson, O.D., David, J., Qi, L.L., et al. 2003a. Synteny perturbations between wheat homoeologous chromosomes caused by locus duplications and deletions correlate with recombination rates. Proc. Natl. Acad. Sci. 100 10836-10841. - PMC - PubMed
-
- Anderson, L.K. and Stack, S.M. 2005. Recombination nodules in plants. Cytogenet. Genome Res. 109 198-204. - PubMed
Web site references
-
- http://www.maizegdb.org; the MaizeGDB database that contains mapping information.
-
- http://www.genome.arizona.edu/; the Arizona Genomics Institute homepage with BAC contig information.
-
- http://rgp.dna.affrc.go.jp/publicdata/geneticmap98/chr10pre.html; Genetic map information for rice chromosome 10 available at the Rice Genome Research Program.
-
- http://www.tigr.org; The Institute for Genomic Research.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
