MAGI-1 is required for Rap1 activation upon cell-cell contact and for enhancement of vascular endothelial cadherin-mediated cell adhesion
- PMID: 16339077
- PMCID: PMC1356604
- DOI: 10.1091/mbc.e05-07-0647
MAGI-1 is required for Rap1 activation upon cell-cell contact and for enhancement of vascular endothelial cadherin-mediated cell adhesion
Abstract
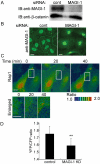
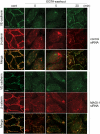
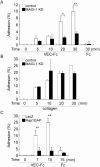
Rap1 is a small GTPase that regulates adherens junction maturation. It remains elusive how Rap1 is activated upon cell-cell contact. We demonstrate for the first time that Rap1 is activated upon homophilic engagement of vascular endothelial cadherin (VE-cadherin) at the cell-cell contacts in living cells and that MAGI-1 is required for VE-cadherin-dependent Rap1 activation. We found that MAGI-1 localized to cell-cell contacts presumably by associating with beta-catenin and that MAGI-1 bound to a guanine nucleotide exchange factor for Rap1, PDZ-GEF1. Depletion of MAGI-1 suppressed the cell-cell contact-induced Rap1 activation and the VE-cadherin-mediated cell-cell adhesion after Ca2+ switch. In addition, relocation of vinculin from cell-extracellular matrix contacts to cell-cell contacts after the Ca2+ switch was inhibited in MAGI-1-depleted cells. Furthermore, inactivation of Rap1 by overexpression of Rap1GAPII impaired the VE-cadherin-dependent cell adhesion. Collectively, MAGI-1 is important for VE-cadherin-dependent Rap1 activation upon cell-cell contact. In addition, once activated, Rap1 upon cell-cell contacts positively regulate the adherens junction formation by relocating vinculin that supports VE-cadherin-based cell adhesion.
Figures





Similar articles
-
Cyclic AMP potentiates vascular endothelial cadherin-mediated cell-cell contact to enhance endothelial barrier function through an Epac-Rap1 signaling pathway.Mol Cell Biol. 2005 Jan;25(1):136-46. doi: 10.1128/MCB.25.1.136-146.2005. Mol Cell Biol. 2005. PMID: 15601837 Free PMC article.
-
Vascular endothelial cadherin-mediated cell-cell adhesion regulated by a small GTPase, Rap1.J Biochem Mol Biol. 2006 Mar 31;39(2):132-9. doi: 10.5483/bmbrep.2006.39.2.132. J Biochem Mol Biol. 2006. PMID: 16584626 Review.
-
Vascular endothelial-cadherin stabilizes at cell-cell junctions by anchoring to circumferential actin bundles through alpha- and beta-catenins in cyclic AMP-Epac-Rap1 signal-activated endothelial cells.Mol Biol Cell. 2010 Feb 15;21(4):584-96. doi: 10.1091/mbc.e09-07-0580. Epub 2009 Dec 23. Mol Biol Cell. 2010. PMID: 20032304 Free PMC article.
-
E-cadherin dis-engagement activates the Rap1 GTPase.J Cell Biochem. 2008 Nov 1;105(4):1027-37. doi: 10.1002/jcb.21902. J Cell Biochem. 2008. PMID: 18767072 Free PMC article.
-
Dynamic Regulation of Vascular Permeability by Vascular Endothelial Cadherin-Mediated Endothelial Cell-Cell Junctions.J Nippon Med Sch. 2017;84(4):148-159. doi: 10.1272/jnms.84.148. J Nippon Med Sch. 2017. PMID: 28978894 Review.
Cited by
-
Polarization of migrating cortical neurons by Rap1 and N-cadherin: Revisiting the model for the Reelin signaling pathway.Small GTPases. 2011 Nov 1;2(6):322-328. doi: 10.4161/sgtp.18283. Small GTPases. 2011. PMID: 22545231 Free PMC article.
-
Up-regulation of the homophilic adhesion molecule sidekick-1 in podocytes contributes to glomerulosclerosis.J Biol Chem. 2010 Aug 13;285(33):25677-85. doi: 10.1074/jbc.M110.133959. Epub 2010 Jun 18. J Biol Chem. 2010. PMID: 20562105 Free PMC article.
-
The small GTPase Rap1 is a novel regulator of RPE cell barrier function.Invest Ophthalmol Vis Sci. 2011 Sep 27;52(10):7455-63. doi: 10.1167/iovs.11-7295. Invest Ophthalmol Vis Sci. 2011. PMID: 21873678 Free PMC article.
-
A mechanism of Rap1-induced stabilization of endothelial cell--cell junctions.Mol Biol Cell. 2011 Jul 15;22(14):2509-19. doi: 10.1091/mbc.E11-02-0157. Epub 2011 Jun 1. Mol Biol Cell. 2011. PMID: 21633110 Free PMC article.
-
Dynamic interplay between adhesion surfaces in carcinomas: Cell-cell and cell-matrix crosstalk.World J Biol Chem. 2016 Feb 26;7(1):64-77. doi: 10.4331/wjbc.v7.i1.64. World J Biol Chem. 2016. PMID: 26981196 Free PMC article. Review.
References
-
- Adamsky, K., Arnold, K., Sabanay, H., and Peles, E. (2003). Junctional protein MAGI-3 interacts with receptor tyrosine phosphatase beta (RPTP beta) and tyrosine-phosphorylated proteins. J. Cell Sci. 116, 1279-1289. - PubMed
-
- Bos, J. L. (2005). Linking Rap to cell adhesion. Curr. Opin. Cell Biol. 17, 123-128. - PubMed
-
- Bos, J. L., de Rooij, J., and Reedquist, K. A. (2001). Rap1 signalling: adhering to new models. Nat. Rev. Mol. Cell Biol. 2, 369-377. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

