Construction of a 2-Mb resolution BAC microarray for CGH analysis of canine tumors
- PMID: 16339382
- PMCID: PMC1356122
- DOI: 10.1101/gr.3825705
Construction of a 2-Mb resolution BAC microarray for CGH analysis of canine tumors
Abstract
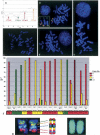
Recognition of the domestic dog as a model for the comparative study of human genetic traits has led to major advances in canine genomics. The pathophysiological similarities shared between many human and dog diseases extend to a range of cancers. Human tumors frequently display recurrent chromosome aberrations, many of which are hallmarks of particular tumor subtypes. Using a range of molecular cytogenetic techniques we have generated evidence indicating that this is also true of canine tumors. Detailed knowledge of these genomic abnormalities has the potential to aid diagnosis, prognosis, and the selection of appropriate therapy in both species. We recently improved the efficiency and resolution of canine cancer cytogenetics studies by developing a small-scale genomic microarray comprising a panel of canine BAC clones representing subgenomic regions of particular interest. We have now extended these studies to generate a comprehensive canine comparative genomic hybridization (CGH) array that comprises 1158 canine BAC clones ordered throughout the genome with an average interval of 2 Mb. Most of the clones (84.3%) have been assigned to a precise cytogenetic location by fluorescence in situ hybridization (FISH), and 98.5% are also directly anchored within the current canine genome assembly, permitting direct translation from cytogenetic aberration to DNA sequence. We are now using this resource routinely for high-throughput array CGH and single-locus probe analysis of a range of canine cancers. Here we provide examples of the varied applications of this resource to tumor cytogenetics, in combination with other molecular cytogenetic techniques.
Figures

References
-
- Bannasch, D.L., Bannasch, M.J., Ryun, J.R., Famula, T.R., and Pedersen, N.C. 2005. Y chromosome haplotype analysis in purebred dogs. Mamm. Genome 16: 273-280. - PubMed
-
- Breen, M., Thomas, R., Faircloth, S., Mahoney, J., Pettengill, M., Scott, A., Thomson, S., Hudson, R., Bianco, S., Fosmire, S., et al. 2003. Molecular cytogenetics of canine lymphoma and leukaemia. In Genes, dogs and cancer: 3rd Annual Canine Cancer Conference. (ed. J. Modiano), pp. P3021.0903. International Veterinary Information Service, Ithaca NY (www.ivis.org), Seattle, WA.
-
- Chung, Y.J., Jonkers, J., Kitson, H., Fiegler, H., Humphray, S., Scott, C., Hunt, S., Yu, Y., Nishijima, I., Velds, A., et al. 2004. A whole-genome mouse BAC microarray with 1-Mb resolution for analysis of DNA copy number changes by array comparative genomic hybridization. Genome Res. 14: 188-196. - PMC - PubMed
Web site references
-
- http://www.cvm.ncsu.edu/mbs/breen_matthew.htm; Primary authors' laboratory Web site, with details of all clones used on this array.
-
- http://cgap.nci.nih.gov/Chromosomes/Mitelman; Mitelman Database of Chromosome Aberrations in Cancer 2005. Mitelman, F., Johansson, B., and Mertens, F. (eds).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
