Envoy, a PAS/LOV domain protein of Hypocrea jecorina (Anamorph Trichoderma reesei), modulates cellulase gene transcription in response to light
- PMID: 16339718
- PMCID: PMC1317494
- DOI: 10.1128/EC.4.12.1998-2007.2005
Envoy, a PAS/LOV domain protein of Hypocrea jecorina (Anamorph Trichoderma reesei), modulates cellulase gene transcription in response to light
Abstract
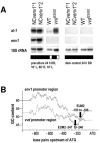
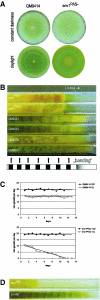
Envoy, a PAS/LOV domain protein with similarity to the Neurospora light regulator Vivid, which has been cloned due to its lack of expression in a cellulase-negative mutant, links cellulase induction by cellulose to light signaling in Hypocrea jecorina. Despite their similarity, env1 could not compensate for the lack of vvd function. Besides the effect of light on sporulation, we observed a reduced growth rate in constant light. An env1(PAS-) mutant of H. jecorina grows significantly slower in the presence of light but remains unaffected in darkness compared to the wild-type strain QM9414. env1 rapidly responds to a light pulse, with this response being different upon growth on glucose or glycerol, and it encodes a regulator essential for H. jecorina light tolerance. The induction of cellulase transcription in H. jecorina by cellulose is enhanced by light in the wild-type strain QM9414 compared to that in constant darkness, whereas a delayed induction in light and only a transient up-regulation in constant darkness of cbh1 was observed in the env1(PAS-) mutant. However, light does not lead to cellulase expression in the absence of an inducer. We conclude that Envoy connects the light response to carbon source signaling and thus that light must be considered an additional external factor influencing gene expression analysis in this fungus.
Figures




References
-
- Ambra, R., B. Grimaldi, S. Zamboni, P. Filetici, G. Macino, and P. Ballario. 2004. Photomorphogenesis in the hypogeous fungus Tuber borchii: isolation and characterization of Tbwc-1, the homologue of the blue-light photoreceptor of Neurospora crassa. Fungal Genet. Biol. 41:688-697. - PubMed
-
- Aro, N., A. Saloheimo, M. Ilmen, and M. Penttila. 2001. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 276:24309-24314. - PubMed
-
- Arpaia, G., F. Cerri, S. Baima, and G. Macino. 1999. Involvement of protein kinase C in the response of Neurospora crassa to blue light. Mol. Gen. Genet. 262:314-322. - PubMed
-
- Ballario, P., and G. Macino. 1997. White collar proteins: PASsing the light signal in Neurospora crassa. Trends Microbiol. 5:458-462. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

