Enzymes of the heme biosynthetic pathway in the nonphotosynthetic alga Polytomella sp
- PMID: 16339726
- PMCID: PMC1317499
- DOI: 10.1128/EC.4.12.2087-2097.2005
Enzymes of the heme biosynthetic pathway in the nonphotosynthetic alga Polytomella sp
Abstract
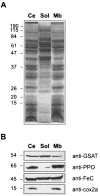
Heme biosynthesis involves a number of enzymatic steps which in eukaryotes take place in different cell compartments. Enzyme compartmentalization differs between photosynthetic and nonphotosynthetic eukaryotes. Here we investigated the structures and subcellular localizations of three enzymes involved in the heme pathway in Polytomella sp., a colorless alga evolutionarily related to the green alga Chlamydomonas reinhardtii. Functional complementation of Escherichia coli mutant strains was used to isolate cDNAs encoding three heme biosynthetic enzymes, glutamate-1-semialdehyde aminotransferase, protoporphyrinogen IX oxidase, and ferrochelatase. All three proteins show highest similarity to their counterparts in photosynthetic organisms, including C. reinhardtii. All three proteins have N-terminal extensions suggestive of intracellular targeting, and immunoblot studies indicate their enrichment in a dense cell fraction that is enriched in amyloplasts. These results suggest that even though the plastids of Polytomella sp. are not photosynthetically active, they are the major site of heme biosynthesis. The presence of a gene for glutamate-1-semialdehyde aminotransferase suggests that Polytomella sp. uses the five-carbon pathway for synthesis of the heme precursor 5-aminolevulinic acid.
Figures




Similar articles
-
Subcellular localization and light-regulated expression of protoporphyrinogen IX oxidase and ferrochelatase in Chlamydomonas reinhardtii.Plant Physiol. 2005 Dec;139(4):1946-58. doi: 10.1104/pp.105.069732. Epub 2005 Nov 23. Plant Physiol. 2005. PMID: 16306143 Free PMC article.
-
The Chlamydomonas reinhardtii gtr gene encoding the tetrapyrrole biosynthetic enzyme glutamyl-trna reductase: structure of the gene and properties of the expressed enzyme.Plant Mol Biol. 2005 Jul;58(5):643-58. doi: 10.1007/s11103-005-6803-x. Plant Mol Biol. 2005. PMID: 16158240
-
Expression and characterization of the terminal heme synthetic enzymes from the hyperthermophile Aquifex aeolicus.FEMS Microbiol Lett. 2001 Aug 7;202(1):115-9. doi: 10.1111/j.1574-6968.2001.tb10789.x. FEMS Microbiol Lett. 2001. PMID: 11506917
-
Glutamyl-transfer RNA: a precursor of heme and chlorophyll biosynthesis.Trends Biochem Sci. 1992 Jun;17(6):215-8. doi: 10.1016/0968-0004(92)90380-r. Trends Biochem Sci. 1992. PMID: 1502723 Review.
-
Structure and function of ferrochelatase.J Bioenerg Biomembr. 1995 Apr;27(2):221-9. doi: 10.1007/BF02110037. J Bioenerg Biomembr. 1995. PMID: 7592569 Review.
Cited by
-
New insights into the unique structure of the F0F1-ATP synthase from the chlamydomonad algae Polytomella sp. and Chlamydomonas reinhardtii.Plant Physiol. 2007 Jun;144(2):1190-9. doi: 10.1104/pp.106.094060. Epub 2007 Apr 27. Plant Physiol. 2007. PMID: 17468226 Free PMC article.
-
A plastid without a genome: evidence from the nonphotosynthetic green algal genus Polytomella.Plant Physiol. 2014 Apr;164(4):1812-9. doi: 10.1104/pp.113.233718. Epub 2014 Feb 21. Plant Physiol. 2014. PMID: 24563281 Free PMC article.
-
Comparative analysis of nucleus-encoded plastid-targeting proteins in Rafflesia cantleyi against photosynthetic and non-photosynthetic representatives reveals orthologous systems with potentially divergent functions.Sci Rep. 2018 Nov 22;8(1):17258. doi: 10.1038/s41598-018-35173-1. Sci Rep. 2018. PMID: 30467394 Free PMC article.
-
Reductive evolution of chloroplasts in non-photosynthetic plants, algae and protists.Curr Genet. 2018 Apr;64(2):365-387. doi: 10.1007/s00294-017-0761-0. Epub 2017 Oct 12. Curr Genet. 2018. PMID: 29026976 Review.
References
-
- Antaramian, A., R. Coria, J. Ramírez, and D. González-Halphen. 1996. The deduced primary structure of subunit I from cytochrome c oxidase suggests that the genus Polytomella shares a common mitochondrial origin with Chlamydomonas. Biochim. Biophys. Acta 1273:198-202. - PubMed
-
- Antaramian, A., S. Funes, M. Vásquez-Acevedo, A. Atteia, R. Coria, and D. González-Halphen. 1998. Two unusual amino acid substitutions in cytochrome b of the colorless alga Polytomella spp.: correlation with the atypical spectral properties of the bH heme. Arch. Biochem. Biophys. 354:206-214. - PubMed
-
- Atteia, A., R. van Lis, J. Ramirez, and D. González-Halphen. 2000. Polytomella spp. growth on ethanol. Extracellular pH affects the accumulation of mitochondrial cytochrome c550. Eur. J. Biochem. 267:2850-2858. - PubMed
-
- Atteia, A., R. van Lis, G. Mendoza-Hernández, K. Henze, W. Martin, H. Riveros-Rosas, and D. González-Halphen. 2003. Bifunctional aldehyde/alcohol dehydrogenase (ADHE) in chlorophyte algal mitochondria. Plant Mol. Biol. 53:175-188. - PubMed
-
- Beale, S. I. 1999. Enzymes of chlorophyll biosynthesis. Photosynth. Res. 60:43-73.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

