RNA-binding domain proteins in Kinetoplastids: a comparative analysis
- PMID: 16339728
- PMCID: PMC1317496
- DOI: 10.1128/EC.4.12.2106-2114.2005
RNA-binding domain proteins in Kinetoplastids: a comparative analysis
Abstract
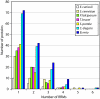
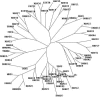
RNA-binding proteins are important in many aspects of RNA processing, function, and destruction. One class of such proteins contains the RNA recognition motif (RRM), which consists of about 90 amino acid residues, including the canonical RNP1 octapeptide: (K/R)G(F/Y)(G/A)FVX(F/Y). We used a variety of homology searches to classify all of the RRM proteins of the three kinetoplastids Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major. All three organisms have similar sets of RRM-containing protein orthologues, suggesting common posttranscriptional processing and regulatory pathways. Of the 75 RRM proteins identified in T. brucei, only 13 had clear homologues in other eukaryotes, although 8 more could be given putative functional assignments. A comparison with the 18 RRM proteins of the obligate intracellular parasite Encephalitozoon cuniculi revealed just 3 RRM proteins which appear to be conserved at the primary sequence level throughout eukaryotic evolution: poly(A) binding protein, the rRNA-processing protein MRD1, and the nuclear cap binding protein.
Figures


References
-
- Batista, J. A. N., S. M. R. Teixeira, J. E. Donelson, L. V. Kirchhoff, and C. Martins de Sá. 1994. Characterization of Trypanosoma cruzi poly(A)-binding protein and its genes. Mol. Biochem. Parasitol. 67:301-312. - PubMed
-
- Benz, C., D. Nilsson, B. Andersson, C. Clayton, and D. L. Guilbride. 2005. Messenger RNA processing sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 143:125-134. - PubMed
-
- Berriman, M., et. al. 2005. The genome of the African trypanosome, Trypanosoma brucei. Science 309:416-422. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

