Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa
- PMID: 16339731
- PMCID: PMC1317490
- DOI: 10.1128/EC.4.12.2140-2152.2005
Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa
Abstract
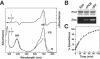
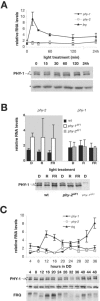
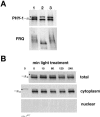
Phytochromes (Phys) comprise a superfamily of red-/far-red-light-sensing proteins. Whereas higher-plant Phys that control numerous growth and developmental processes have been well described, the biochemical characteristics and functions of the microbial forms are largely unknown. Here, we describe analyses of the expression, regulation, and activities of two Phys in the filamentous fungus Neurospora crassa. In addition to containing the signature N-terminal domain predicted to covalently associate with a bilin chromophore, PHY-1 and PHY-2 contain C-terminal histidine kinase and response regulator motifs, implying that they function as hybrid two-component sensor kinases activated by light. A bacterially expressed N-terminal fragment of PHY-2 covalently bound either biliverdin or phycocyanobilin in vitro, with the resulting holoprotein displaying red-/far-red-light photochromic absorption spectra and a photocycle in vitro. cDNA analysis of phy-1 and phy-2 revealed two splice isoforms for each gene. The levels of the phy transcripts are not regulated by light, but the abundance of the phy-1 mRNAs is under the control of the circadian clock. Phosphorylated and unphosphorylated forms of PHY-1 were detected; both species were found exclusively in the cytoplasm, with their relative abundances unaffected by light. Strains containing deletions of phy-1 and phy-2, either singly or in tandem, were not compromised in any known photoresponses in Neurospora, leaving their function(s) unclear.
Figures
Similar articles
-
Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors.Biochem J. 2005 Nov 15;392(Pt 1):103-16. doi: 10.1042/BJ20050826. Biochem J. 2005. PMID: 16004604 Free PMC article.
-
The Fast-Evolving phy-2 Gene Modulates Sexual Development in Response to Light in the Model Fungus Neurospora crassa.mBio. 2016 Mar 8;7(2):e02148. doi: 10.1128/mBio.02148-15. mBio. 2016. PMID: 26956589 Free PMC article.
-
Phytochrome controls conidiation in response to red/far-red light and daylight length and regulates multistress tolerance in Beauveria bassiana.Environ Microbiol. 2014 Jul;16(7):2316-28. doi: 10.1111/1462-2920.12486. Epub 2014 May 5. Environ Microbiol. 2014. PMID: 24725588
-
White collar proteins: PASsing the light signal in Neurospora crassa.Trends Microbiol. 1997 Nov;5(11):458-62. doi: 10.1016/S0966-842X(97)01144-X. Trends Microbiol. 1997. PMID: 9402704 Review.
-
The phytochromes: a biochemical mechanism of signaling in sight?Bioessays. 1997 Jul;19(7):571-9. doi: 10.1002/bies.950190708. Bioessays. 1997. PMID: 9230690 Review.
Cited by
-
Streptophyte phytochromes exhibit an N-terminus of cyanobacterial origin and a C-terminus of proteobacterial origin.BMC Res Notes. 2015 Apr 13;8:144. doi: 10.1186/s13104-015-1082-3. BMC Res Notes. 2015. PMID: 25886068 Free PMC article.
-
Genome-wide characterization of light-regulated genes in Neurospora crassa.G3 (Bethesda). 2014 Jul 21;4(9):1731-45. doi: 10.1534/g3.114.012617. G3 (Bethesda). 2014. PMID: 25053707 Free PMC article.
-
The fungal pathogen Aspergillus fumigatus regulates growth, metabolism, and stress resistance in response to light.mBio. 2013 Mar 26;4(2):e00142-13. doi: 10.1128/mBio.00142-13. mBio. 2013. PMID: 23532976 Free PMC article.
-
Regulation of conidiation by light in Aspergillus nidulans.Genetics. 2011 Aug;188(4):809-22. doi: 10.1534/genetics.111.130096. Epub 2011 May 30. Genetics. 2011. PMID: 21624998 Free PMC article.
-
The Neurospora crassa DCC-1 protein, a putative histidine kinase, is required for normal sexual and asexual development and carotenogenesis.Eukaryot Cell. 2011 Dec;10(12):1733-9. doi: 10.1128/EC.05223-11. Epub 2011 Nov 4. Eukaryot Cell. 2011. PMID: 22058142 Free PMC article.
References
-
- Aronson, B. D., K. A. Johnson, J. J. Loros, and J. C. Dunlap. 1994. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science 263:1578-1584. - PubMed
-
- Baima, S., G. Macino, and G. Morelli. 1991. Photoregulation of the albino-3 gene in Neurospora crassa. J. Photochem. Photobiol. B 11:107-115. - PubMed
-
- Bhoo, S. H., S. J. Davis, J. Walker, B. Karniol, and R. D. Vierstra. 2001. Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature 414:776-779. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

