Alternative sigma factors and their roles in bacterial virulence
- PMID: 16339734
- PMCID: PMC1306804
- DOI: 10.1128/MMBR.69.4.527-543.2005
Alternative sigma factors and their roles in bacterial virulence
Abstract
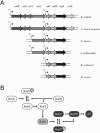
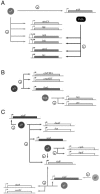
Sigma factors provide promoter recognition specificity to RNA polymerase holoenzyme, contribute to DNA strand separation, and then dissociate from the core enzyme following transcription initiation. As the regulon of a single sigma factor can be composed of hundreds of genes, sigma factors can provide effective mechanisms for simultaneously regulating expression of large numbers of prokaryotic genes. One newly emerging field is identification of the specific roles of alternative sigma factors in regulating expression of virulence genes and virulence-associated genes in bacterial pathogens. Virulence genes encode proteins whose functions are essential for the bacterium to effectively establish an infection in a host organism. In contrast, virulence-associated genes can contribute to bacterial survival in the environment and therefore may enhance the capacity of the bacterium to spread to new individuals or to survive passage through a host organism. As alternative sigma factors have been shown to regulate expression of both virulence and virulence-associated genes, these proteins can contribute both directly and indirectly to bacterial virulence. Sigma factors are classified into two structurally unrelated families, the sigma70 and the sigma54 families. The sigma70 family includes primary sigma factors (e.g., Bacillus subtilis sigma(A)) as well as related alternative sigma factors; sigma54 forms a distinct subfamily of sigma factors referred to as sigma(N) in almost all species for which these proteins have been characterized to date. We present several examples of alternative sigma factors that have been shown to contribute to virulence in at least one organism. For each sigma factor, when applicable, examples are drawn from multiple species.
Figures
References
-
- Alarcón-Chaidez, F. J., L. Keith, Y. Zhao, and C. L. Bender. 2003. RpoN (σ54) is required for plasmid-encoded coronatine biosynthesis in Pseudomonas syringae. Plasmid 49:106-117. - PubMed
-
- Bashyam, M. D., and S. E. Hasnain. 2004. The extracytoplasmic function sigma factors: role in bacterial pathogenesis. Infect. Genet. Evol. 4:301-308. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

