The 2-hydroxycarboxylate transporter family: physiology, structure, and mechanism
- PMID: 16339740
- PMCID: PMC1306803
- DOI: 10.1128/MMBR.69.4.665-695.2005
The 2-hydroxycarboxylate transporter family: physiology, structure, and mechanism
Abstract
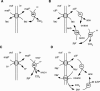
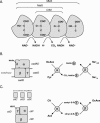
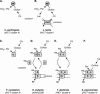
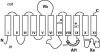
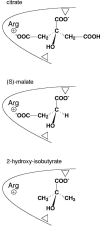
The 2-hydroxycarboxylate transporter family is a family of secondary transporters found exclusively in the bacterial kingdom. They function in the metabolism of the di- and tricarboxylates malate and citrate, mostly in fermentative pathways involving decarboxylation of malate or oxaloacetate. These pathways are found in the class Bacillales of the low-CG gram-positive bacteria and in the gamma subdivision of the Proteobacteria. The pathways have evolved into a remarkable diversity in terms of the combinations of enzymes and transporters that built the pathways and of energy conservation mechanisms. The transporter family includes H+ and Na+ symporters and precursor/product exchangers. The proteins consist of a bundle of 11 transmembrane helices formed from two homologous domains containing five transmembrane segments each, plus one additional segment at the N terminus. The two domains have opposite orientations in the membrane and contain a pore-loop or reentrant loop structure between the fourth and fifth transmembrane segments. The two pore-loops enter the membrane from opposite sides and are believed to be part of the translocation site. The binding site is located asymmetrically in the membrane, close to the interface of membrane and cytoplasm. The binding site in the translocation pore is believed to be alternatively exposed to the internal and external media. The proposed structure of the 2HCT transporters is different from any known structure of a membrane protein and represents a new structural class of secondary transporters.
Figures


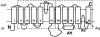

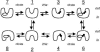

References
-
- Abe, K., Z. S. Ruan, and P. C. Maloney. 1996. Cloning, sequencing, and expression in Escherichia coli of OxlT, the oxalate:formate exchange protein of Oxalobacter formigens. J. Biol. Chem. 271:6789-6793. - PubMed
-
- Abramson, J., I. Smirnova, V. Kasho, G. Verner, H. R. Kaback, and S. Iwata. 2003. Structure and mechanism of the lactose permease of Escherichia coli. Science 301:610-615. - PubMed
-
- Accardi, A., and C. Miller. 2004. Secondary active transport mediated by a prokaryotic homologue of ClC Cl− channels. Nature 427:803-807. - PubMed
-
- Anantharam, V., M. J. Allison, and P. C. Maloney. 1989. Oxalate:formate exchange. The basis for energy coupling in Oxalobacter. J. Biol. Chem. 264:7244-7250. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

