Structure and architecture of the maize genome
- PMID: 16339807
- PMCID: PMC1310546
- DOI: 10.1104/pp.105.068718
Structure and architecture of the maize genome
Abstract
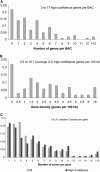
Maize (Zea mays or corn) plays many varied and important roles in society. It is not only an important experimental model plant, but also a major livestock feed crop and a significant source of industrial products such as sweeteners and ethanol. In this study we report the systematic analysis of contiguous sequences of the maize genome. We selected 100 random regions averaging 144 kb in size, representing about 0.6% of the genome, and generated a high-quality dataset for sequence analysis. This sampling contains 330 annotated genes, 91% of which are supported by expressed sequence tag data from maize and other cereal species. Genes averaged 4 kb in size with five exons, although the largest was over 59 kb with 31 exons. Gene density varied over a wide range from 0.5 to 10.7 genes per 100 kb and genes did not appear to cluster significantly. The total repetitive element content we observed (66%) was slightly higher than previous whole-genome estimates (58%-63%) and consisted almost exclusively of retroelements. The vast majority of genes can be aligned to at least one sequence read derived from gene-enrichment procedures, but only about 30% are fully covered. Our results indicate that much of the increase in genome size of maize relative to rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana) is attributable to an increase in number of both repetitive elements and genes.
Figures



References
-
- Alleman M, Doctor J (2000) Genomic imprinting in plants: observations and evolutionary implications. Plant Mol Biol 43: 147–161 - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 - PubMed
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

