A papain-like enzyme at work: native and acyl-enzyme intermediate structures in phytochelatin synthesis
- PMID: 16339904
- PMCID: PMC1310510
- DOI: 10.1073/pnas.0505833102
A papain-like enzyme at work: native and acyl-enzyme intermediate structures in phytochelatin synthesis
Abstract
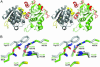
Phytochelatin synthase (PCS) is a key enzyme for heavy-metal detoxification in plants. PCS catalyzes the production of glutathione (GSH)-derived peptides (called phytochelatins or PCs) that bind heavy-metal ions before vacuolar sequestration. The enzyme can also hydrolyze GSH and GS-conjugated xenobiotics. In the cyanobacterium Nostoc, the enzyme (NsPCS) contains only the catalytic domain of the eukaryotic synthase and can act as a GSH hydrolase and weakly as a peptide ligase. The crystal structure of NsPCS in its native form solved at a 2.0-A resolution shows that NsPCS is a dimer that belongs to the papain superfamily of cysteine proteases, with a conserved catalytic machinery. Moreover, the structure of the protein solved as a complex with GSH at a 1.4-A resolution reveals a gamma-glutamyl cysteine acyl-enzyme intermediate stabilized in a cavity of the protein adjacent to a second putative GSH binding site. GSH hydrolase and PCS activities of the enzyme are discussed in the light of both structures.
Figures
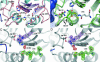

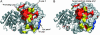
Comment in
-
Phytochelatin synthase, papain's cousin, in stereo.Proc Natl Acad Sci U S A. 2006 Jan 17;103(3):507-8. doi: 10.1073/pnas.0509971102. Epub 2006 Jan 9. Proc Natl Acad Sci U S A. 2006. PMID: 16407124 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

