Interaction of nephrocystin-4 and RPGRIP1 is disrupted by nephronophthisis or Leber congenital amaurosis-associated mutations
- PMID: 16339905
- PMCID: PMC1317916
- DOI: 10.1073/pnas.0505774102
Interaction of nephrocystin-4 and RPGRIP1 is disrupted by nephronophthisis or Leber congenital amaurosis-associated mutations
Abstract
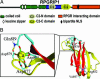
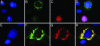
RPGR-interacting protein 1 (RPGRIP1) is a key component of cone and rod photoreceptor cells, where it interacts with RPGR (retinitis pigmentosa GTPase regulator). Mutations in RPGRIP1 lead to autosomal recessive congenital blindness [Leber congenital amaurosis (LCA)]. Most LCA-associated missense mutations in RPGRIP1 are located in a segment that encodes two C2 domains. Based on the C2 domain of novel protein kinase C epsilon (PKC epsilon), we built a 3D-homology model for the C-terminal C2 domain of RPGRIP1. This model revealed a potential Ca2+-binding site that was predicted to be disrupted by a missense mutation in RPGRIP1, which was previously identified in an LCA patient. Through yeast two-hybrid screening of a retinal cDNA library, we found this C2 domain to specifically bind to nephrocystin-4, encoded by NPHP4. Mutations in NPHP4 are associated with nephronophthisis and a combination of nephronophthisis and retinitis pigmentosa called Senior-Løken syndrome (SLSN). We show that RPGRIP1 and nephrocystin-4 interact strongly in vitro and in vivo, and that they colocalize in the retina, matching the panretinal localization pattern of specific RPGRIP1 isoforms. Their interaction is disrupted by either mutations in RPGRIP1, found in patients with LCA, or by mutations in NPHP4, found in patients with nephronophthisis or SLSN. Thus, we provide evidence for the involvement of this disrupted interaction in the retinal dystrophy of both SLSN and LCA patients.
Figures
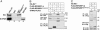


Similar articles
-
Selective loss of RPGRIP1-dependent ciliary targeting of NPHP4, RPGR and SDCCAG8 underlies the degeneration of photoreceptor neurons.Cell Death Dis. 2012 Jul 19;3(7):e355. doi: 10.1038/cddis.2012.96. Cell Death Dis. 2012. PMID: 22825473 Free PMC article.
-
RPGRIP1 is mutated in Leber congenital amaurosis: a mini-review.Ophthalmic Genet. 2005 Dec;26(4):175-9. doi: 10.1080/13816810500374441. Ophthalmic Genet. 2005. PMID: 16352478 Review.
-
Limited proteolysis differentially modulates the stability and subcellular localization of domains of RPGRIP1 that are distinctly affected by mutations in Leber's congenital amaurosis.Hum Mol Genet. 2005 May 15;14(10):1327-40. doi: 10.1093/hmg/ddi143. Epub 2005 Mar 30. Hum Mol Genet. 2005. PMID: 15800011 Free PMC article.
-
The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors.Hum Mol Genet. 2000 Sep 1;9(14):2095-105. doi: 10.1093/hmg/9.14.2095. Hum Mol Genet. 2000. PMID: 10958648
-
Leber congenital amaurosis caused by mutations in RPGRIP1.Cold Spring Harb Perspect Med. 2014 Nov 20;5(4):a017384. doi: 10.1101/cshperspect.a017384. Cold Spring Harb Perspect Med. 2014. PMID: 25414380 Free PMC article. Review.
Cited by
-
Advances in the understanding of nuclear pore complexes in human diseases.J Cancer Res Clin Oncol. 2024 Jul 30;150(7):374. doi: 10.1007/s00432-024-05881-5. J Cancer Res Clin Oncol. 2024. PMID: 39080077 Free PMC article. Review.
-
Selective loss of RPGRIP1-dependent ciliary targeting of NPHP4, RPGR and SDCCAG8 underlies the degeneration of photoreceptor neurons.Cell Death Dis. 2012 Jul 19;3(7):e355. doi: 10.1038/cddis.2012.96. Cell Death Dis. 2012. PMID: 22825473 Free PMC article.
-
Evidence for RPGRIP1 gene as risk factor for primary open angle glaucoma.Eur J Hum Genet. 2011 Apr;19(4):445-51. doi: 10.1038/ejhg.2010.217. Epub 2011 Jan 12. Eur J Hum Genet. 2011. PMID: 21224891 Free PMC article.
-
Mutations in CTNNA1 cause butterfly-shaped pigment dystrophy and perturbed retinal pigment epithelium integrity.Nat Genet. 2016 Feb;48(2):144-51. doi: 10.1038/ng.3474. Epub 2015 Dec 21. Nat Genet. 2016. PMID: 26691986 Free PMC article.
-
The primary cilium as a cellular signaling center: lessons from disease.Curr Opin Genet Dev. 2009 Jun;19(3):220-9. doi: 10.1016/j.gde.2009.04.008. Epub 2009 May 22. Curr Opin Genet Dev. 2009. PMID: 19477114 Free PMC article. Review.
References
-
- Roepman, R., van Duynhoven, G., Rosenberg, T., Pinckers, A. J. L. G., Bleeker-Wagemakers, E. M., Bergen, A. A. B., Post, J., Beck, A., Reinhardt, R., Ropers, H.-H., et al. (1996) Hum. Mol. Genet. 5, 1035-1041. - PubMed
-
- Meindl, A., Dry, K., Herrmann, K., Manson, F., Ciccodicola, A., Edgar, A., Carvalho, M. R., Achatz, H., Hellebrand, H., Lennon, A., et al. (1996) Nat. Genet. 13, 35-42. - PubMed
-
- Yang, Z., Peachey, N. S., Moshfeghi, D. M., Thirumalaichary, S., Chorich, L., Shugart, Y. Y., Fan, K. & Zhang, K. (2002) Hum. Mol. Genet. 11, 605-611. - PubMed
-
- Ayyagari, R., Demirci, F., Liu, J., Bingham, E., Stringham, H., Kakuk, L., Boehnke, M., Gorin, M., Richards, J. & Sieving, P. (2002) Genomics 80, 166-171. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

