The evolutionary dynamics of alpha-satellite
- PMID: 16344556
- PMCID: PMC1356132
- DOI: 10.1101/gr.3810906
The evolutionary dynamics of alpha-satellite
Abstract
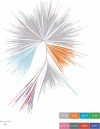
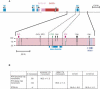
Alpha-satellite is a family of tandemly repeated sequences found at all normal human centromeres. In addition to its significance for understanding centromere function, alpha-satellite is also a model for concerted evolution, as alpha-satellite repeats are more similar within a species than between species. There are two types of alpha-satellite in the human genome; while both are made up of approximately 171-bp monomers, they can be distinguished by whether monomers are arranged in extremely homogeneous higher-order, multimeric repeat units or exist as more divergent monomeric alpha-satellite that lacks any multimeric periodicity. In this study, as a model to examine the genomic and evolutionary relationships between these two types, we have focused on the chromosome 17 centromeric region that has reached both higher-order and monomeric alpha-satellite in the human genome assembly. Monomeric and higher-order alpha-satellites on chromosome 17 are phylogenetically distinct, consistent with a model in which higher-order evolved independently of monomeric alpha-satellite. Comparative analysis between human chromosome 17 and the orthologous chimpanzee chromosome indicates that monomeric alpha-satellite is evolving at approximately the same rate as the adjacent non-alpha-satellite DNA. However, higher-order alpha-satellite is less conserved, suggesting different evolutionary rates for the two types of alpha-satellite.
Figures
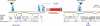


References
-
- Alexandrov, I.A., Mashkova, T.D., Akopian, T.A., Medvedev, L.I., Kisselev, L.L., Mitkevich, S.P., and Yurov, Y.B. 1991. Chromosome-specific α satellites: Two distinct families on human chromosome 18. Genomics 11 15-23. - PubMed
-
- Alexandrov, I., Kazakov, A., Tumeneva, I., Shepelev, V., and Yurov, Y. 2001. α-Satellite DNA of primates: Old and new families. Chromosoma 110 253-266. - PubMed
-
- Alkan, C., Eichler, E.E., Bailey, J.A., Sahinalp, S.C., and Tuzun, E. 2004. The role of unequal crossover in α-satellite DNA evolution: A computational analysis. J. Comp. Biol. 11 933-944. - PubMed
-
- Alves, G., Seuanez, H.N., and Fanning, T. 1994. α-Satellite DNA in neotropical primates (Platyrrhini). Chromosoma 103 262-267. - PubMed
Web site references
-
- http://darwin.uvigo.es/software/modeltest.html; Modeltest.
-
- http://genome.ucsc.edu; UCSC Genome Bioinformatics site.
-
- http://macclade.org/macclade.html; MacClade.
-
- http://pipeline.lbl.gov/cgi-bin/gateway2; VISTA browser.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
