Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome
- PMID: 16344566
- PMCID: PMC1356123
- DOI: 10.1101/gr.4222606
Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome
Abstract
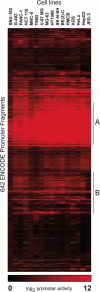
Transcriptional promoters comprise one of many classes of eukaryotic transcriptional regulatory elements. Identification and characterization of these elements are vital to understanding the complex network of human gene regulation. Using full-length cDNA sequences to identify transcription start sites (TSS), we predicted more than 900 putative human transcriptional promoters in the ENCODE regions, representing a comprehensive sampling of promoters in 1% of the genome. We identified 387 fragments that function as promoters in at least one of 16 cell lines by measuring promoter activity in high-throughput transient transfection reporter assays. These positive functional results demonstrate widespread use of alternative promoters. We show a strong correlation between promoter activity and the corresponding endogenous RNA transcript levels, providing the first experimental quantitative estimate of promoter contribution to gene regulation. Finally, we identified functional regions within a randomly selected subset of 45 promoters using deletion analyses. These experiments showed that, on average, the sequence -300 to -50 bp of the TSS positively contributes to core promoter activity. Interestingly, putative negative elements were identified -1000 to -500 bp upstream of the TSS for 55% of genes tested. These data provide the largest and most comprehensive view of promoter function in the human genome.
Figures




References
-
- Ahituv, N., Rubin, E.M., and Nobrega, M.A. 2004. Exploiting human-fish genome comparisons for deciphering gene regulation. Hum. Mol. Genet. 13 Spec No 2: R261-R266. - PubMed
-
- Bailey, T.L. and Elkan, C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2 28-36. - PubMed
-
- Bertone, P., Stolc, V., Royce, T.E., Rozowsky, J.S., Urban, A.E., Zhu, X., Rinn, J.L., Tongprasit, W., Samanta, M., Weissman, S., et al. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306 2242-2246. - PubMed
-
- Blais, A. and Dynlacht, B.D. 2004. Hitting their targets: An emerging picture of E2F and cell cycle control. Curr. Opin. Genet. Dev. 14 527-532. - PubMed
Web site references
-
- http://genome.ucsc.edu/ENCODE; ENCODE.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
