The Histone Database: a comprehensive resource for histones and histone fold-containing proteins
- PMID: 16345076
- PMCID: PMC1800941
- DOI: 10.1002/prot.20814
The Histone Database: a comprehensive resource for histones and histone fold-containing proteins
Abstract
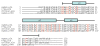
The Histone Database is a curated and searchable collection of full-length sequences and structures of histones and nonhistone proteins containing histone-like folds, compiled from major public databases. Several new histone fold-containing proteins have been identified, including the huntingtin-interacting protein HYPM. Additionally, based on the recent crystal structure of the Son of Sevenless protein, an interpretation of the sequence analysis of the histone fold domain is presented. The database contains an updated collection of multiple sequence alignments for the four core histones (H2A, H2B, H3, and H4) and the linker histones (H1/H5) from a total of 975 organisms. The database also contains information on the human histone gene complement and provides links to three-dimensional structures of histone and histone fold-containing proteins. The Histone Database is a comprehensive bioinformatics resource for the study of structure and function of histones and histone fold-containing proteins. The database is available at http://research.nhgri.nih.gov/histones/.
2005 Wiley-Liss, Inc.
Figures


References
-
- van Holde KE. Chromatin. New York: Springer-Verlag; 1988.
-
- Eickbush TH, Moudrianakis EN. The histone core complex: an octamer assembled by two sets of protein-protein interactions. Biochemistry. 1978;17(23):4955– 4964. - PubMed
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389(6648):251–260. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

