Purification and Characterization of Exo-beta-d-Glucosaminidase from a Cellulolytic Fungus, Trichoderma reesei PC-3-7
- PMID: 16349528
- PMCID: PMC106342
- DOI: 10.1128/AEM.64.3.890-895.1998
Purification and Characterization of Exo-beta-d-Glucosaminidase from a Cellulolytic Fungus, Trichoderma reesei PC-3-7
Abstract
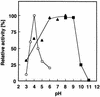
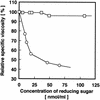
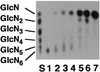
Chitosan-degrading activities induced by glucosamine (GlcN) or N-acetylglucosamine (GlcNAc) were found in a culture filtrate of Trichoderma reesei PC-3-7. One of the chitosan-degrading enzymes was purified to homogeneity by precipitation with ammonium sulfate followed by anion-exchange and hydrophobic-interaction chromatographies. The enzyme was monomeric, and its molecular mass was 93 kDa. The optimum pH and temperature of the enzyme were 4.0 and 50 degrees C, respectively. The activity was stable in the pH range 6.0 to 9.0 and at a temperature below 50 degrees C. Reaction product analysis from the viscosimetric assay and thin-layer chromatography and H nuclear magnetic resonance spectroscopy clearly indicated that the enzyme was an exo-type chitosanase, exo-beta-d-glucosaminidase, that releases GlcN from the nonreducing end of the chitosan chain. H nuclear magnetic resonance spectroscopy also showed that the exo-beta-d-glucosaminidase produced a beta-form of GlcN, demonstrating that the enzyme is a retaining glycanase. Time-dependent liberation of the reducing sugar from partially acetylated chitosan with exo-beta-d-glucosaminidase and the partially purified exo-beta-d-N-acetylglucosaminidase from T. reesei PC-3-7 suggested that the exo-beta-d-glucosaminidase cleaves the glycosidic link of either GlcN-beta(1-->4)-GlcN or GlcN-beta(1-->4)-GlcNAc.
Figures




References
-
- Alfonso C, Martinez M J, Reyes F. Purification and properties of two endochitosanases from Mucor rouxii implicated in its cell wall degradation. FEMS Microbiol Lett. 1992;95:187–194.
-
- Bartnicki-Garcia S. Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu Rev Microbiol. 1968;22:87–108. - PubMed
-
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Carmen Limón M, Lora J M, García I, de la Cruz J, Llobell A, Benítez T, Pintor-Toro J A. Primary structure and expression pattern of the 33-kDa chitinase gene from the mycoparasitic fungus Trichoderma harzianum. Curr Genet. 1995;28:478–483. - PubMed
-
- El Ouakfaoui S, Asselin A. Multiple forms of chitosanase activities. Phytochemistry. 1992;31:1513–1518.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

