Characterization of early steps in the poliovirus infection process: receptor-decorated liposomes induce conversion of the virus to membrane-anchored entry-intermediate particles
- PMID: 16352541
- PMCID: PMC1317540
- DOI: 10.1128/JVI.80.1.172-180.2006
Characterization of early steps in the poliovirus infection process: receptor-decorated liposomes induce conversion of the virus to membrane-anchored entry-intermediate particles
Abstract
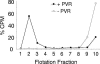
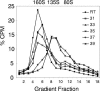
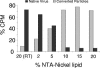
The mechanism by which poliovirus infects the cell has been characterized by a combination of biochemical and structural studies, leading to a working model for cell entry. Upon receptor binding at physiological temperature, native virus (160S) undergoes a conformational change to a 135S particle from which VP4 and the N terminus of VP1 are externalized. These components interact with the membrane and are proposed to form a membrane pore. An additional conformational change in the particle is accompanied by release of the infectious viral RNA genome from the particle and its delivery, presumably through the membrane pore into the cytoplasm, leaving behind an empty 80S particle. In this report, we describe the generation of a receptor-decorated liposome system, comprising nickel-chelating nitrilotriacetic acid (NTA) liposomes and His-tagged poliovirus receptor, and its use in characterizing the early events in poliovirus infection. Receptor-decorated liposomes were able to capture virus and induce a temperature-dependent virus conversion to the 135S particle. Upon conversion, 135S particles became tethered to the liposome independently of receptor by a membrane interaction with the N terminus of VP1. Converted particles had lost VP4, which partitioned with the membrane. The development of a simple model membrane system provides a novel tool for studying poliovirus entry. The liposome system bridges the gap between previous studies using either soluble receptor or whole cells and offers a flexible template which can be extrapolated to electron microscopy experiments that analyze the structural biology of nonenveloped virus entry.
Figures






Similar articles
-
A mutation in VP4 defines a new step in the late stages of cell entry by poliovirus.J Virol. 1993 Aug;67(8):5075-8. doi: 10.1128/JVI.67.8.5075-5078.1993. J Virol. 1993. PMID: 8392631 Free PMC article.
-
Is the 135S poliovirus particle an intermediate during cell entry?J Virol. 2000 Sep;74(18):8757-61. doi: 10.1128/jvi.74.18.8757-8761.2000. J Virol. 2000. PMID: 10954579 Free PMC article.
-
Real-Time Imaging of Polioviral RNA Translocation across a Membrane.mBio. 2021 Feb 23;12(1):e03695-20. doi: 10.1128/mBio.03695-20. mBio. 2021. PMID: 33622727 Free PMC article.
-
Poliovirus and apoptosis.Prog Mol Subcell Biol. 2004;36:151-69. doi: 10.1007/978-3-540-74264-7_8. Prog Mol Subcell Biol. 2004. PMID: 15171611 Review. No abstract available.
-
Poliovirus cell entry: common structural themes in viral cell entry pathways.Annu Rev Microbiol. 2002;56:677-702. doi: 10.1146/annurev.micro.56.012302.160757. Epub 2002 Jan 30. Annu Rev Microbiol. 2002. PMID: 12142481 Free PMC article. Review.
Cited by
-
Cryo-EM structures reveal two distinct conformational states in a picornavirus cell entry intermediate.PLoS Pathog. 2020 Sep 30;16(9):e1008920. doi: 10.1371/journal.ppat.1008920. eCollection 2020 Sep. PLoS Pathog. 2020. PMID: 32997730 Free PMC article.
-
Why Cells and Viruses Cannot Survive without an ESCRT.Cells. 2021 Feb 24;10(3):483. doi: 10.3390/cells10030483. Cells. 2021. PMID: 33668191 Free PMC article. Review.
-
Intraspecies host specificity of a single-stranded RNA virus infecting a marine photosynthetic protist is determined at the early steps of infection.J Virol. 2007 Feb;81(3):1372-8. doi: 10.1128/JVI.01082-06. Epub 2006 Nov 15. J Virol. 2007. PMID: 17108022 Free PMC article.
-
Backbone Free Energy Estimator Applied to Viral Glycoproteins.J Comput Biol. 2020 Oct;27(10):1495-1508. doi: 10.1089/cmb.2020.0120. Epub 2020 Apr 3. J Comput Biol. 2020. PMID: 32250657 Free PMC article.
-
Unraveling the Motions behind Enterovirus 71 Uncoating.Biophys J. 2018 Feb 27;114(4):822-838. doi: 10.1016/j.bpj.2017.12.021. Biophys J. 2018. PMID: 29490244 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

