Defective assembly of influenza A virus due to a mutation in the polymerase subunit PA
- PMID: 16352550
- PMCID: PMC1317532
- DOI: 10.1128/JVI.80.1.252-261.2006
Defective assembly of influenza A virus due to a mutation in the polymerase subunit PA
Abstract
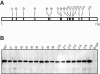
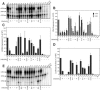
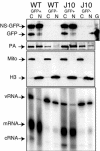
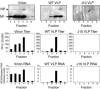
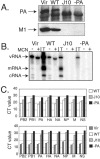
The RNA-dependent RNA polymerase of influenza A virus is composed of three subunits that together synthesize all viral mRNAs and also replicate the viral genomic RNA segments (vRNAs) through intermediates known as cRNAs. Here we describe functional characterization of 16 site-directed mutants of one polymerase subunit, termed PA. In accord with earlier studies, these mutants exhibited diverse, mainly quantitative impairments in expressing one or more classes of viral RNA, with associated infectivity defects of varying severity. One PA mutant, however, targeting residues 507 and 508, caused only modest perturbations of RNA expression yet completely eliminated the formation of plaque-forming virus. Polymerases incorporating this mutant, designated J10, proved capable of synthesizing translationally active mRNAs and of replicating diverse cRNA or vRNA templates at levels compatible with viral infectivity. Both the mutant protein and its RNA products were appropriately localized in the cytoplasm, where influenza virus assembly occurs. Nevertheless, J10 failed to generate infectious particles from cells in a plasmid-based influenza virus assembly assay, and hemagglutinating material from the supernatants of such cells contained little or no nuclease-resistant genomic RNA. These findings suggest that PA has a previously unrecognized role in assembly or release of influenza virus virions, perhaps influencing core structure or the packaging of vRNAs or other essential components into nascent influenza virus particles.
Figures

Similar articles
-
Mutational analyses of the influenza A virus polymerase subunit PA reveal distinct functions related and unrelated to RNA polymerase activity.PLoS One. 2012;7(1):e29485. doi: 10.1371/journal.pone.0029485. Epub 2012 Jan 6. PLoS One. 2012. PMID: 22238617 Free PMC article.
-
Segment-specific noncoding sequences of the influenza virus genome RNA are involved in the specific competition between defective interfering RNA and its progenitor RNA segment at the virion assembly step.J Virol. 1997 Mar;71(3):2138-45. doi: 10.1128/JVI.71.3.2138-2145.1997. J Virol. 1997. PMID: 9032347 Free PMC article.
-
The molecular anatomy of influenza virus RNA polymerase.Biol Chem. 1997 Jun;378(6):483-8. Biol Chem. 1997. PMID: 9224927 Review.
-
Threonine 157 of influenza virus PA polymerase subunit modulates RNA replication in infectious viruses.J Virol. 2003 May;77(10):6007-13. doi: 10.1128/jvi.77.10.6007-6013.2003. J Virol. 2003. PMID: 12719592 Free PMC article.
-
[Structural and functional aspects of influenza virus RNA polymerase].Zhonghua Jie He He Hu Xi Za Zhi. 2004 Apr;27(4):261-3. Zhonghua Jie He He Hu Xi Za Zhi. 2004. PMID: 15144619 Review. Chinese. No abstract available.
Cited by
-
Conserved features of the PB2 627 domain impact influenza virus polymerase function and replication.J Virol. 2014 Jun;88(11):5977-86. doi: 10.1128/JVI.00508-14. Epub 2014 Mar 12. J Virol. 2014. PMID: 24623411 Free PMC article.
-
Generation and Comprehensive Analysis of Host Cell Interactome of the PA Protein of the Highly Pathogenic H5N1 Avian Influenza Virus in Mammalian Cells.Front Microbiol. 2017 Apr 28;8:739. doi: 10.3389/fmicb.2017.00739. eCollection 2017. Front Microbiol. 2017. PMID: 28503168 Free PMC article.
-
Influenza virus recruits host protein kinase C to control assembly and activity of its replication machinery.Elife. 2017 Jul 31;6:e26910. doi: 10.7554/eLife.26910. Elife. 2017. PMID: 28758638 Free PMC article.
-
Detection and characterization of influenza A virus PA-PB2 interaction through a bimolecular fluorescence complementation assay.J Virol. 2009 Apr;83(8):3944-55. doi: 10.1128/JVI.02300-08. Epub 2009 Feb 4. J Virol. 2009. PMID: 19193801 Free PMC article.
-
Influenza A virus polymerase is a site for adaptive changes during experimental evolution in bat cells.J Virol. 2014 Nov;88(21):12572-85. doi: 10.1128/JVI.01857-14. Epub 2014 Aug 20. J Virol. 2014. PMID: 25142579 Free PMC article.
References
-
- Delaluna, S., C. Martinez, and J. Ortin. 1989. Molecular cloning and sequencing of influenza virus A/Victoria/3/75 polymerase genes—sequence evolution and prediction of possible functional domains. Virus Res. 13:143-156. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

