Characterization of mimivirus DNA topoisomerase IB suggests horizontal gene transfer between eukaryal viruses and bacteria
- PMID: 16352556
- PMCID: PMC1317558
- DOI: 10.1128/JVI.80.1.314-321.2006
Characterization of mimivirus DNA topoisomerase IB suggests horizontal gene transfer between eukaryal viruses and bacteria
Abstract
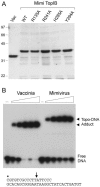
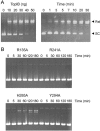
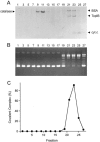
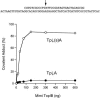
Mimivirus, a parasite of Acanthamoeba polyphaga, is the largest DNA virus known; it encodes dozens of proteins with imputed functions in nucleic acid transactions. Here we produced, purified, and characterized mimivirus DNA topoisomerase IB (TopIB), which we find to be a structural and functional homolog of poxvirus TopIB and the poxvirus-like topoisomerases discovered recently in bacteria. Arginine, histidine, and tyrosine side chains responsible for TopIB transesterification are conserved and essential in mimivirus TopIB. Moreover, mimivirus TopIB is capable of incising duplex DNA at the 5'-CCCTT cleavage site recognized by all poxvirus topoisomerases. Based on the available data, mimivirus TopIB appears functionally more akin to poxvirus TopIB than bacterial TopIB, despite its greater primary structure similarity to the bacterial TopIB group. We speculate that the ancestral bacterial/viral TopIB was disseminated by horizontal gene transfer within amoebae, which are permissive hosts for either intracellular growth or persistence of many present-day bacterial species that have a type IB topoisomerase.
Figures


References
-
- Baylis, S. A., L. K. Dixon, S. Vydelingum, and G. L. Smith. 1992. African swine fever virus encodes a gene with extensive homology to type II DNA topoisomerases. J. Mol. Biol. 228:1003-1010. - PubMed
-
- Cheng, C., L. K. Wang, J. Sekiguchi, and S. Shuman. 1997. Mutational analysis of 39 residues of vaccinia DNA topoisomerase identifies Lys-220, Arg-223, and Asn-228 as important for covalent catalysis. J. Biol. Chem. 272:8263-8269. - PubMed
-
- Cheng, C., P. Kussie, N. Pavletich, and S. Shuman. 1998. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell 92:841-850. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

