Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase
- PMID: 16352565
- PMCID: PMC1317551
- DOI: 10.1128/JVI.80.1.404-411.2006
Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase
Abstract
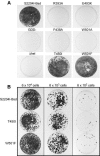
Mutations were introduced into the NS3 helicase region of a hepatitis C virus (HCV) Con1 subgenomic replicon to ascertain the role of the helicase in viral replication. One new replicon lacked two-thirds of the NS3 helicase (Deltahel), and six others contained one of the following six amino acid substitutions in NS3: R393A, F438A, T450I, E493K, W501A, and W501F. It has been previously reported that purified R393A, F438A, and W501A HCV helicase proteins do not unwind RNA but unwind DNA, bind RNA, and hydrolyze ATP. On the other hand, previous data suggest that a W501F protein retains most of its unwinding abilities and that purified T450I and E493K HCV helicase proteins have enhanced unwinding abilities. In a hepatoma cell line that has been cured of HCV replicons using interferon, the T450I and W501F replicons synthesized both negative-sense and positive-sense viral RNA and formed colonies after selection with similar efficiencies as the parent replicon. However, the Deltahel, R393A, F438A, and W501A replicons encoded and processed an HCV polyprotein but did not synthesize additional viral RNA or form colonies. Surprisingly the same phenotype was seen for the E493K replicon. The inability of the E493K replicon to replicate might point to a role of pH in viral replication because a previous analysis has shown that, unlike the wild-type NS3 protein, the helicase activity of an E493K protein is not sensitive to pH changes. These results demonstrate that the RNA-unwinding activity of the HCV NS3 helicase is needed for RNA replication.
Figures

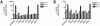

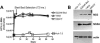
References
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. - PubMed
-
- Cho, H. S., N. C. Ha, L. W. Kang, K. M. Chung, S. H. Back, S. K. Jang, and B. H. Oh. 1998. Crystal structure of RNA helicase from genotype 1b hepatitis C virus. A feasible mechanism of unwinding duplex RNA. J. Biol. Chem. 273:15045-15052. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

