Nonstructural protein 3 of bluetongue virus assists virus release by recruiting ESCRT-I protein Tsg101
- PMID: 16352570
- PMCID: PMC1317520
- DOI: 10.1128/JVI.80.1.460-473.2006
Nonstructural protein 3 of bluetongue virus assists virus release by recruiting ESCRT-I protein Tsg101
Abstract
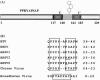
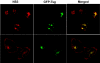
The release of Bluetongue virus (BTV) and other members of the Orbivirus genus from infected host cells occurs predominantly by cell lysis, and in some cases, by budding from the plasma membrane. Two nonstructural proteins, NS3 and NS3A, have been implicated in this process. Here we show that both proteins bind to human Tsg101 and its ortholog from Drosophila melanogaster with similar strengths in vitro. This interaction is mediated by a conserved PSAP motif in NS3 and appears to play a role in virus release. The depletion of Tsg101 with small interfering RNA inhibits the release of BTV and African horse sickness virus, a related orbivirus, from HeLa cells up to fivefold and threefold, respectively. Like most other viral proteins which recruit Tsg101, NS3 also harbors a PPXY late-domain motif that allows NS3 to bind NEDD4-like ubiquitin ligases in vitro. However, the late-domain motifs in NS3 do not function as effectively in facilitating the release of mini Gag virus-like particles from 293T cells as the late domains from human immunodeficiency virus type 1, human T-cell leukemia virus, and Ebola virus. A mutagenesis study showed that the arginine residue in the PPRY motif is responsible for the low activity of the NS3 late-domain motifs. Our data suggest that the BTV late-domain motifs either recruit an antagonist that interferes with budding or fail to recruit an agonist which is different from NEDD4.
Figures





Similar articles
-
A viral nonstructural protein regulates bluetongue virus trafficking and release.J Virol. 2009 Jul;83(13):6806-16. doi: 10.1128/JVI.00263-09. Epub 2009 Apr 15. J Virol. 2009. PMID: 19369335 Free PMC article.
-
PPPYVEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101 [corrected].J Virol. 2003 Nov;77(22):11882-95. doi: 10.1128/jvi.77.22.11882-11895.2003. J Virol. 2003. PMID: 14581525 Free PMC article.
-
Bluetongue Virus Nonstructural Protein 3 Orchestrates Virus Maturation and Drives Non-Lytic Egress via Two Polybasic Motifs.Viruses. 2019 Nov 29;11(12):1107. doi: 10.3390/v11121107. Viruses. 2019. PMID: 31795485 Free PMC article.
-
Novel Tsg101 Binding Partners Regulate Viral L Domain Trafficking.Viruses. 2021 Jun 15;13(6):1147. doi: 10.3390/v13061147. Viruses. 2021. PMID: 34203832 Free PMC article. Review.
-
[HIV budding and Tsg101].Uirusu. 2005 Dec;55(2):281-6. doi: 10.2222/jsv.55.281. Uirusu. 2005. PMID: 16557014 Review. Japanese.
Cited by
-
Vaccination as a Strategy to Prevent Bluetongue Virus Vertical Transmission.Pathogens. 2021 Nov 22;10(11):1528. doi: 10.3390/pathogens10111528. Pathogens. 2021. PMID: 34832683 Free PMC article. Review.
-
Bluetongue virus assembly and exit pathways.Adv Virus Res. 2020;108:249-273. doi: 10.1016/bs.aivir.2020.08.002. Epub 2020 Sep 16. Adv Virus Res. 2020. PMID: 33837718 Free PMC article. Review.
-
Bluetongue virus outer capsid protein VP5 interacts with membrane lipid rafts via a SNARE domain.J Virol. 2008 Nov;82(21):10600-12. doi: 10.1128/JVI.01274-08. Epub 2008 Aug 27. J Virol. 2008. PMID: 18753209 Free PMC article.
-
Identification and characterization of host cell proteins interacting with Scylla serrata reovirus non-structural protein p35.Virus Genes. 2017 Apr;53(2):317-322. doi: 10.1007/s11262-016-1418-7. Epub 2016 Dec 9. Virus Genes. 2017. PMID: 27943061
-
Viruses exploit the tissue physiology of the host to spread in vivo.Curr Opin Cell Biol. 2016 Aug;41:81-90. doi: 10.1016/j.ceb.2016.04.008. Epub 2016 May 3. Curr Opin Cell Biol. 2016. PMID: 27149407 Free PMC article. Review.
References
-
- Babst, M., D. J. Katzmann, E. J. Estepa-Sabal, T. Meerloo, and S. D. Emr. 2002. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3:271-282. - PubMed
-
- Babst, M., D. J. Katzmann, W. B. Snyder, B. Wendland, and S. D. Emr. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3:283-289. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

