Crimean-congo hemorrhagic fever virus glycoprotein precursor is cleaved by Furin-like and SKI-1 proteases to generate a novel 38-kilodalton glycoprotein
- PMID: 16352575
- PMCID: PMC1317557
- DOI: 10.1128/JVI.80.1.514-525.2006
Crimean-congo hemorrhagic fever virus glycoprotein precursor is cleaved by Furin-like and SKI-1 proteases to generate a novel 38-kilodalton glycoprotein
Abstract
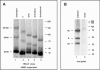
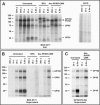
Crimean-Congo hemorrhagic fever virus (genus Nairovirus, family Bunyaviridae) genome M segment encodes an unusually large (in comparison to members of other genera) polyprotein (1,684 amino acids in length) containing the two major structural glycoproteins, Gn and Gc, that are posttranslationally processed from precursors PreGn and PreGc by SKI-1 and SKI-1-like proteases, respectively. The characteristics of the N-terminal 519 amino acids located upstream of the mature Gn are unknown. A highly conserved furin/proprotein convertase (PC) cleavage site motif (RSKR247) is located between the variable N-terminal region that is predicted to have mucin-like properties and the rest of PreGn. Mutational analysis of the RSKR247 motif and use of a specific furin/PC inhibitor and brefeldin A demonstrate that furin/PC cleavage occurs at the RSKR247 motif of PreGn as the protein transits the trans Golgi network and generates a novel glycoprotein designated GP38. Immunoprecipitation analysis identified two additional proteins, GP85 and GP160, which contain both mucin and GP38 domain regions, and whose generation does not involve furin/PC cleavage. Consistent with glycosylation predictions, heavy O-linked glycosylation and moderate levels of N-glycans were detected in the GP85 and GP160 proteins, both of which contain the mucin domain. GP38, GP85, and GP160 are likely soluble proteins based on the lack of predicted transmembrane domains, their detection in virus-infected cell supernatants, and the apparent absence from virions. Analogy with soluble glycoproteins and mucin-like proteins encoded by other hemorrhagic fever-associated RNA viruses suggests these proteins could play an important role in viral pathogenesis.
Figures


Similar articles
-
Recovery of Recombinant Crimean Congo Hemorrhagic Fever Virus Reveals a Function for Non-structural Glycoproteins Cleavage by Furin.PLoS Pathog. 2015 May 1;11(5):e1004879. doi: 10.1371/journal.ppat.1004879. eCollection 2015 May. PLoS Pathog. 2015. PMID: 25933376 Free PMC article.
-
Crimean-Congo hemorrhagic fever virus glycoprotein proteolytic processing by subtilase SKI-1.J Virol. 2003 Aug;77(16):8640-9. doi: 10.1128/jvi.77.16.8640-8649.2003. J Virol. 2003. PMID: 12885882 Free PMC article.
-
Identification of a novel C-terminal cleavage of Crimean-Congo hemorrhagic fever virus PreGN that leads to generation of an NSM protein.J Virol. 2007 Jun;81(12):6632-42. doi: 10.1128/JVI.02730-06. Epub 2007 Apr 4. J Virol. 2007. PMID: 17409136 Free PMC article.
-
How Do Enveloped Viruses Exploit the Secretory Proprotein Convertases to Regulate Infectivity and Spread?Viruses. 2021 Jun 25;13(7):1229. doi: 10.3390/v13071229. Viruses. 2021. PMID: 34202098 Free PMC article. Review.
-
Structure and function of eukaryotic proprotein processing enzymes of the subtilisin family of serine proteases.Crit Rev Oncog. 1993;4(2):115-36. Crit Rev Oncog. 1993. PMID: 8420571 Review.
Cited by
-
Crimean-Congo hemorrhagic fever virus glycoprotein processing by the endoprotease SKI-1/S1P is critical for virus infectivity.J Virol. 2007 Dec;81(23):13271-6. doi: 10.1128/JVI.01647-07. Epub 2007 Sep 26. J Virol. 2007. PMID: 17898072 Free PMC article.
-
Basolateral entry and release of Crimean-Congo hemorrhagic fever virus in polarized MDCK-1 cells.J Virol. 2007 Mar;81(5):2158-64. doi: 10.1128/JVI.02070-06. Epub 2006 Dec 13. J Virol. 2007. PMID: 17166898 Free PMC article.
-
Structure and Characterization of Crimean-Congo Hemorrhagic Fever Virus GP38.J Virol. 2020 Mar 31;94(8):e02005-19. doi: 10.1128/JVI.02005-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31996434 Free PMC article.
-
Long-lived CD8+ T cell responses following Crimean-Congo haemorrhagic fever virus infection.PLoS Negl Trop Dis. 2017 Dec 19;11(12):e0006149. doi: 10.1371/journal.pntd.0006149. eCollection 2017 Dec. PLoS Negl Trop Dis. 2017. PMID: 29261651 Free PMC article.
-
Molecular Detection and Genetic Characterization of Two Dugbe Orthonairovirus Isolates Detected from Ticks in Southern Senegal.Viruses. 2024 Jun 15;16(6):964. doi: 10.3390/v16060964. Viruses. 2024. PMID: 38932256 Free PMC article.
References
-
- Barr, P. J. 1991. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell 66:1-3. - PubMed
-
- Basak, A., M. Zhong, J. S. Munzer, M. Chrétien, and N. G. Seidah. 2001. Implication of the proprotein convertases furin, PC5 and PC7 in the cleavage of surface glycoproteins of Hong Kong, Ebola and respiratory syncytial viruses: a comparative analysis with fluorogenic peptides. Biochem. J. 353:537-545. - PMC - PubMed
-
- Bertolotti-Ciarlet, A., J. Smith, K. Strecker, J. Paragas, L. A. Altamura, J. M. McFalls, N. Frias-Stäheli, A. García-Sastre, C. S. Schmaljohn, and R. W. Doms. 2005. Cellular localization and antigenic characterization of Crimean-Congo hemorrhagic fever virus glycoproteins. J. Virol. 79:6152-6161. - PMC - PubMed
-
- Buchmeier, M. J., M. D. Bowen, and C. J. Peters. 2001. Arenaviridae: the viruses and their replication, p. 1635-1668. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 2nd ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

