Two types of precursor cells in a multipotential hematopoietic cell line
- PMID: 16352715
- PMCID: PMC1317970
- DOI: 10.1073/pnas.0509314102
Two types of precursor cells in a multipotential hematopoietic cell line
Abstract
The biochemistry of early stages of hematopoietic differentiation is difficult to study because only relatively small numbers of precursor cells are available. The murine EML cell line is a multipotential cell line that can be used to model some of these steps. We found that the lineage- EML precursor cells can be separated into two populations based on cell surface markers including CD34. Both populations contain similar levels of stem cell factor (SCF) receptor (c-Kit) but only the CD34+ population shows a growth response when treated with SCF. Conversely, the CD34- population will grow in the presence of the cytokine IL-3. The human beta-globin locus control region hypersensitive site 2 plays different roles on beta-globin transcription in the CD34+ and CD34- populations. The two populations are present in about equal amounts in culture, and the CD34+ population rapidly regenerates the mixed population when grown in the presence of SCF. We suggest that this system may mimic a normal developmental transition in hematopoiesis.
Figures
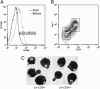

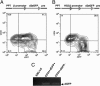



Similar articles
-
Tcf7 is an important regulator of the switch of self-renewal and differentiation in a multipotential hematopoietic cell line.PLoS Genet. 2012;8(3):e1002565. doi: 10.1371/journal.pgen.1002565. Epub 2012 Mar 8. PLoS Genet. 2012. PMID: 22412390 Free PMC article.
-
Complex interactions in EML cell stimulation by stem cell factor and IL-3.Proc Natl Acad Sci U S A. 2011 Mar 22;108(12):4882-7. doi: 10.1073/pnas.1018002108. Epub 2011 Mar 7. Proc Natl Acad Sci U S A. 2011. PMID: 21383156 Free PMC article.
-
c-kit is expressed by primitive human hematopoietic cells that give rise to colony-forming cells in stroma-dependent or cytokine-supplemented culture.Exp Hematol. 1994 Feb;22(2):157-65. Exp Hematol. 1994. PMID: 7507857
-
Stem cell factor as a survival and growth factor in human normal and malignant hematopoiesis.Acta Haematol. 1996;95(3-4):257-62. doi: 10.1159/000203893. Acta Haematol. 1996. PMID: 8677752 Review.
-
Regulation of mast cell development.Chem Immunol Allergy. 2005;87:1-21. doi: 10.1159/000087566. Chem Immunol Allergy. 2005. PMID: 16107759 Review.
Cited by
-
Tcf7 is an important regulator of the switch of self-renewal and differentiation in a multipotential hematopoietic cell line.PLoS Genet. 2012;8(3):e1002565. doi: 10.1371/journal.pgen.1002565. Epub 2012 Mar 8. PLoS Genet. 2012. PMID: 22412390 Free PMC article.
-
Identification of key factors regulating self-renewal and differentiation in EML hematopoietic precursor cells by RNA-sequencing analysis.J Vis Exp. 2014 Nov 11;(93):e52104. doi: 10.3791/52104. J Vis Exp. 2014. PMID: 25407807 Free PMC article.
-
A CO-FISH assay to assess sister chromatid segregation patterns in mitosis of mouse embryonic stem cells.Chromosome Res. 2013 May;21(3):311-28. doi: 10.1007/s10577-013-9358-8. Chromosome Res. 2013. PMID: 23681662 Free PMC article.
-
Genetic fingerprint defines hematopoietic stem cell pool size and function.Haematologica. 2020 Mar;105(3):526-528. doi: 10.3324/haematol.2019.241299. Haematologica. 2020. PMID: 32115410 Free PMC article. No abstract available.
-
The role of tumor suppressor p15Ink4b in the regulation of hematopoietic progenitor cell fate.Blood Cancer J. 2013 Jan;3(1):e99. doi: 10.1038/bcj.2012.44. Epub 2013 Jan 4. Blood Cancer J. 2013. PMID: 23359317 Free PMC article.
References
-
- Tsai, S., Bartelmez, S., Sitnicka, E. & Collins, S. (1994) Genes Dev. 8, 2831–2841. - PubMed
-
- Yu, W. M., Hawley, T. S., Hawley, R. G. & Qu, C. K. (2002) Blood 100, 3828–3831. - PubMed
-
- Reya, T., Duncan, A. W., Ailles, L., Domen, J., Scherer, D. C., Willert, K., Hintz, L., Nusse, R. & Weissman, I. L. (2003) Nature 423, 409–414. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

