A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element
- PMID: 16352720
- PMCID: PMC1311908
- DOI: 10.1073/pnas.0507693102
A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element
Abstract
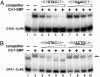
The CRR1 (Copper Response Regulator) locus, required for both activating and repressing target genes of a copper- and hypoxia-sensing pathway in Chlamydomonas, encodes a 1,232-residue candidate transcription factor with a plant-specific DNA-binding domain named SBP, ankyrin repeats, and a C-terminal Cys-rich region, with similarity to a Drosophila metallothionein. The recombinant SBP domain of Crr1 shows zinc-dependent binding to functionally defined copper-response elements associated with the CYC6 and CPX1 promoters that contain a critical GTAC core sequence. Competition experiments indicate equivalent selectivity for copper-response elements from either promoter and 10-fold greater selectivity for the wild-type sequence vs. a sequence carrying a single mutation in the GTAC core. The SBP domain of Chlamydomonas Crr1 binds also to a related GTAC-containing sequence in the Arabidopsis AP1 promoter that is the binding site of a defining member of the SBP family of DNA-binding proteins. Chlamydomonas Crr1 is most similar to a subset of the Arabidopsis SBP domain proteins, which include SPL1, SPL7, and SPL12. The abundance of the CRR1 mRNA is only marginally copper-responsive, and although two mRNAs that differ with respect to splicing of the first intron are detected, there is no indication that the splicing event is regulated by metal nutrition or hypoxia. It is likely that the dramatic copper-responsive action of Crr1 occurs at the level of the polypeptide.
Figures




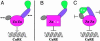
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

