Septum enlightenment: assembly of bacterial division proteins
- PMID: 16352817
- PMCID: PMC1317574
- DOI: 10.1128/JB.188.1.19-27.2006
Septum enlightenment: assembly of bacterial division proteins
Figures

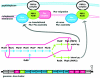
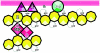

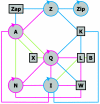
References
-
- Aarsman, M. E., A. Piette, C. Fraipont, T. M. Vinkenvleugel, M. Nguyen-Distéche, and T. den Blaauwen. 2005. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 55:1631-1645. - PubMed
-
- Addinall, S. G., C. Cao, and J. Lutkenhaus. 1997. FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol. 25:303-309. - PubMed
-
- Ayala, J. A., T. Garrido, M. A. de Pedro, and M. Vicente. 1994. Molecular biology of bacterial septation. .In J. M. Ghuysen and R. Habenneck (ed.), Bacterial cell wall. Elsevier Science, Amsterdam, The Netherlands.
-
- Barondess, J. J., M. Carson, L. M. Guzmán Verduzco, and J. Beckwith. 1991. Alkaline phosphatase fusions in the study of cell division genes. Res. Microbiol. 142:295-299. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

