Identification and characterization of the genes involved in glycosylation pathways of mycobacterial glycopeptidolipid biosynthesis
- PMID: 16352824
- PMCID: PMC1317587
- DOI: 10.1128/JB.188.1.86-95.2006
Identification and characterization of the genes involved in glycosylation pathways of mycobacterial glycopeptidolipid biosynthesis
Abstract
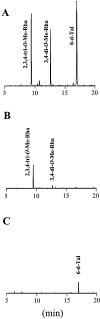
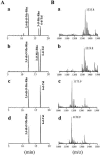
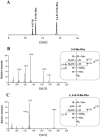
Glycopeptidolipids (GPLs) are major components present on the outer layers of the cell walls of several nontuberculous mycobacteria. GPLs are antigenic molecules and have variant oligosaccharides in mycobacteria such as Mycobacterium avium. In this study, we identified four genes (gtf1, gtf2, gtf3, and gtf4) in the genome of Mycobacterium smegmatis. These genes were independently inactivated by homologous recombination in M. smegmatis, and the structures of GPLs from each gene disruptant were analyzed. Thin-layer chromatography, gas chromatography-mass spectrometry, and matrix-assisted laser desorption ionization-time-of-flight mass spectrometry analyses revealed that the mutants Deltagtf1 and Deltagtf2 accumulated the fatty acyl-tetrapeptide core having O-methyl-rhamnose and 6-deoxy-talose as sugar residues, respectively. The mutant Deltagtf4 possessed the same GPLs as the wild type, whereas the mutant Deltagtf3 lacked two minor GPLs, consisting of 3-O-methyl-rhamnose attached to O-methyl-rhamnose of the fatty acyl-tetrapeptide core. These results indicate that the gtf1 and gtf2 genes are responsible for the early glycosylation steps of GPL biosynthesis and the gtf3 gene is involved in transferring a rhamnose residue not to 6-deoxy-talose but to an O-methyl-rhamnose residue. Moreover, a complementation experiment showed that M. avium gtfA and gtfB, which are deduced glycosyltransferase genes of GPL biosynthesis, restore complete GPL production in the mutants Deltagtf1 and Deltagtf2, respectively. Our findings propose that both M. smegmatis and M. avium have the common glycosylation pathway in the early steps of GPL biosynthesis but differ at the later stages.
Figures



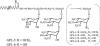


Similar articles
-
Structural characterization of a specific glycopeptidolipid containing a novel N-acyl-deoxy sugar from mycobacterium intracellulare serotype 7 and genetic analysis of its glycosylation pathway.J Bacteriol. 2007 Feb;189(3):1099-108. doi: 10.1128/JB.01471-06. Epub 2006 Nov 22. J Bacteriol. 2007. PMID: 17122347 Free PMC article.
-
The mycobacterial glycopeptidolipids: structure, function, and their role in pathogenesis.Glycobiology. 2008 Nov;18(11):832-41. doi: 10.1093/glycob/cwn076. Epub 2008 Aug 22. Glycobiology. 2008. PMID: 18723691 Free PMC article. Review.
-
Characterization of the fucosylation pathway in the biosynthesis of glycopeptidolipids from Mycobacterium avium complex.J Bacteriol. 2007 Aug;189(15):5515-22. doi: 10.1128/JB.00344-07. Epub 2007 May 25. J Bacteriol. 2007. PMID: 17526707 Free PMC article.
-
Methylation of GPLs in Mycobacterium smegmatis and Mycobacterium avium.J Bacteriol. 2004 Oct;186(20):6792-9. doi: 10.1128/JB.186.20.6792-6799.2004. J Bacteriol. 2004. PMID: 15466031 Free PMC article.
-
Mycobacterium avium and modulation of the host macrophage immune mechanisms.Int J Tuberc Lung Dis. 2011 Apr;15(4):447-52. doi: 10.5588/ijtld.09.0695. Int J Tuberc Lung Dis. 2011. PMID: 21396201 Review.
Cited by
-
Structural characterization of a specific glycopeptidolipid containing a novel N-acyl-deoxy sugar from mycobacterium intracellulare serotype 7 and genetic analysis of its glycosylation pathway.J Bacteriol. 2007 Feb;189(3):1099-108. doi: 10.1128/JB.01471-06. Epub 2006 Nov 22. J Bacteriol. 2007. PMID: 17122347 Free PMC article.
-
A GMC oxidoreductase homologue is required for acetylation of glycopeptidolipid in Mycobacterium smegmatis.Biochemistry. 2014 Feb 4;53(4):611-3. doi: 10.1021/bi4015083. Epub 2014 Jan 24. Biochemistry. 2014. PMID: 24444367 Free PMC article.
-
Genome Analysis and Characterisation of the Exopolysaccharide Produced by Bifidobacterium longum subsp. longum 35624™.PLoS One. 2016 Sep 22;11(9):e0162983. doi: 10.1371/journal.pone.0162983. eCollection 2016. PLoS One. 2016. PMID: 27656878 Free PMC article.
-
Identification and characterization of two novel methyltransferase genes that determine the serotype 12-specific structure of glycopeptidolipids of Mycobacterium intracellulare.J Bacteriol. 2008 Feb;190(3):1064-71. doi: 10.1128/JB.01370-07. Epub 2007 Nov 16. J Bacteriol. 2008. PMID: 18024513 Free PMC article.
-
The mycobacterial glycopeptidolipids: structure, function, and their role in pathogenesis.Glycobiology. 2008 Nov;18(11):832-41. doi: 10.1093/glycob/cwn076. Epub 2008 Aug 22. Glycobiology. 2008. PMID: 18723691 Free PMC article. Review.
References
-
- Aspinall, G. O., D. Chatterjee, and P. J. Brennan. 1995. The variable surface glycolipids of mycobacteria: structures, synthesis of epitopes, and biological properties. Adv. Carbohydr. Chem. Biochem. 51:169-242. - PubMed
-
- Bardarov, S., S. Bardarov, Jr., M. S. Pavelka, Jr., V. Sambandamurthy, M. Larsen, J. Tufariello, J. Chan, G. Hatfull, and W. R. Jacobs, Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG, and M. smegmatis. Microbiology 148:3007-3017. - PubMed
-
- Belisle, J. T., K. Klaczkiewicz, P. J. Brennan, W. R. Jacobs, Jr., and J. M. Inamine. 1993. Rough morphological variants of Mycobacterium avium. Characterization of genomic deletions resulting in the loss of glycopeptidolipid expression. J. Biol. Chem. 268:10517-10523. - PubMed
-
- Billman-Jacobe, H., M. J. McConville, R. E. Haites, S. Kovacevic, and R. L. Coppel. 1999. Identification of a peptide synthetase involved in the biosynthesis of glycopeptidolipids of Mycobacterium smegmatis. Mol. Microbiol. 33:1244-1253. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

