The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili
- PMID: 16352829
- PMCID: PMC1317580
- DOI: 10.1128/JB.188.1.132-140.2006
The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili
Abstract
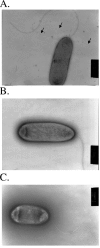
Pseudomonas aeruginosa is an opportunistic pathogen that is commonly found in water and soil. In order to colonize surfaces with low water content, P. aeruginosa utilizes a flagellum-independent form of locomotion called twitching motility, which is dependent upon the extension and retraction of type IV pili. This study demonstrates that AlgZ, previously identified as a DNA-binding protein absolutely required for transcription of the alginate biosynthetic operon, is required for twitching motility. AlgZ may be required for the biogenesis or function of type IV pili in twitching motility. Transmission electron microscopy analysis of an algZ deletion in nonmucoid PAO1 failed to detect surface pili. To examine expression and localization of PilA (the major pilin subunit), whole-cell extracts and cell surface pilin preparations were analyzed by Western blotting. While the PilA levels present in whole-cell extracts were similar for wild-type P. aeruginosa and P. aeruginosa with the algZ deletion, the amount of PilA on the surface of the cells was drastically reduced in the algZ mutant. Analysis of algZ and algD mutants indicates that the DNA-binding activity of AlgZ is essential for the regulation of twitching motility and that this is independent of the role of AlgZ in alginate expression. These data show that AlgZ DNA-binding activity is required for twitching motility independently of its role in alginate production and that this involves the surface localization of type IV pili. Given this new role in twitching motility, we propose that algZ (PA3385) be designated amrZ (alginate and motility regulator Z).
Figures




References
-
- Baynham, P. J., A. L. Brown, L. L. Hall, and D. J. Wozniak. 1999. Pseudomonas aeruginosa AlgZ, a ribbon-helix-helix DNA-binding protein, is essential for alginate synthesis and algD transcriptional activation. Mol. Microbiol. 33:1069-1080. - PubMed
-
- Baynham, P. J., and D. J. Wozniak. 1996. Identification and characterization of AlgZ, an AlgT-dependent DNA-binding protein required for Pseudomonas aeruginosa algD transcription. Mol. Microbiol. 22:97-108. - PubMed
-
- Bradley, D. E. 1980. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can. J. Microbiol. 26:146-154. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

