Transposon disruption of the complex I NADH oxidoreductase gene (snoD) in Staphylococcus aureus is associated with reduced susceptibility to the microbicidal activity of thrombin-induced platelet microbicidal protein 1
- PMID: 16352837
- PMCID: PMC1317573
- DOI: 10.1128/JB.188.1.211-222.2006
Transposon disruption of the complex I NADH oxidoreductase gene (snoD) in Staphylococcus aureus is associated with reduced susceptibility to the microbicidal activity of thrombin-induced platelet microbicidal protein 1
Abstract
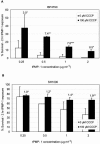
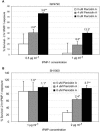
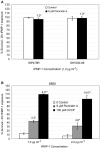
The cationic molecule thrombin-induced platelet microbicidal protein 1 (tPMP-1) exerts potent activity against Staphylococcus aureus. We previously reported that a Tn551 S. aureus transposon mutant, ISP479R, and two bacteriophage back-transductants, TxA and TxB, exhibit reduced in vitro susceptibility to tPMP-1 (tPMP-1(r)) compared to the parental strain, ISP479C (V. Dhawan, M. R. Yeaman, A. L. Cheung, E. Kim, P. M. Sullam, and A. S. Bayer, Infect. Immun. 65:3293-3299, 1997). In the current study, the genetic basis for tPMP-1(r) in these mutants was identified. GenBank homology searches using sequence corresponding to chromosomal DNA flanking Tn551 mutant strains showed that the fourth gene in the staphylococcal mnh operon (mnhABCDEFG) was insertionally inactivated. This operon was previously reported to encode a Na(+)/H(+) antiporter involved in pH tolerance and halotolerance. However, the capacity of ISP479R to grow at pH extremes and in high NaCl concentrations (1 to 3 M), coupled with its loss of transmembrane potential (DeltaPsi) during postexponential growth, suggested that the mnh gene products are not functioning as a secondary (i.e., passive) Na(+)/H(+) antiporter. Moreover, we identified protein homologies between mnhD and the nuo genes of Escherichia coli that encode components of a complex I NADH:ubiquinone oxidoreductase. Consistent with these data, exposures of tPMP-1-susceptible (tPMP-1(s)) parental strains (both clinical and laboratory derived) with either CCCP (a proton ionophore which collapses the proton motive force) or pieracidin A (a specific complex I enzyme inhibitor) significantly reduced tPMP-induced killing to levels seen in the tPMP-1(r) mutants. To reflect the energization of the gene products encoded by the mnh operon, we have renamed the locus sno (S. aureus nuo orthologue). These novel findings indicate that disruption of a complex I enzyme locus can confer reduced in vitro susceptibility to tPMP-1 in S. aureus.
Figures
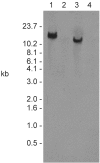
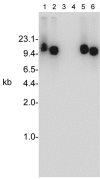



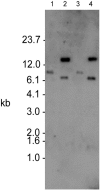
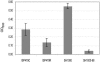


References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Ausubel, F. A., R. Brent, R. E. Kingston, R. E. Moore, D. D. Steidman, J. G. Smith, J. A. Smith, and K. Struhl (ed.). 1996. Current protocols in molecular biology. Greene Publishing and Wiley Interscience, New York, N.Y.
-
- Barquera, B., P. Hellwig, W. Zhou, J. E. Morgan, C. C. Hase, K. K. Gosink, M. Nilges, P. J. Bruesehoff, A. Roth, C. R. Lancaster, and R. B. Gennis. 2002. Purification and characterization of the recombinant Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry 41:3781-3789. - PubMed
-
- Baumert, N., C. von Eiff, F. Schaaff, G. Peters, R. A. Proctor, and H. G. Sahl. 2002. Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb. Drug Resist. 8:253-260. - PubMed
-
- Bayer, A. S., R. P. Prasad, J. Chandra, A. Koul, M. Smriti, A. Varma, R. A. Skurray, N. Firth, M. H. Brown, S.-P. Koo, and M. R. Yeaman. 2000. In vitro resistance of Staphylococcus aureus to thrombin-induced platelet microbicidal protein in associated with alterations in cytoplasmic membrane fluidity. Infect. Immun. 68:3548-3553. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

